1 Aas JA, Paster BJ, Stokes LN, et al. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721.
2 Adamson M, Carlsson J. Lactoperoxidase and thiocyanate protect bacteria from hydrogen peroxide. Infect Immun. 1982;35:20.
3 Adell R, Lekholm U, Rockler B, et al. Marginal tissue reactions at osseointegrated titanium fixtures (I). A 3-year longitudinal prospective study. Int J Oral Maxillofac Surg. 1986;15:39.
4 Adriaens PA, De Boever JA, Loesche WJ. Bacterial invasion in root cementum and radicular dentin of periodontally diseased teeth in humans. A reservoir of periodontopathic bacteria. J Periodontol. 1988;59:222.
5 Ainamo J, Asikainen S, Ainamo A, et al. Plaque growth while chewing sorbitol and xylitol simultaneously with sucrose flavored gum. J Clin Periodontol. 1979;6:397.
6 Alaluusua S, Renkonen OV. Streptococcus mutans establishment and dental caries experience in children from 2 to 4 years old. Scand J Dent Res. 1983;91:453.
7 Alcoforado GA, Slots J. Actinobacillus actinomycetemcomitans and black-pigmented bacteroides in advanced periodontitis in man. Theoretical and practical considerations. Rev Port Estomatol Cir Maxilofac. 1990;31:89.
8 Allison DG, Gilbert P. Modification by surface association of antimicrobial susceptibility of bacterial populations. J Ind Microbiol. 1995;15:311.
9 Alpagot T, Wolff LF, Smith QT, et al. Risk indicators for periodontal disease in a racially diverse urban population. J Clin Periodontol. 1996;23:982.
10 Amano A, Nakagawa I, Okahashi N, et al. Variations of Porphyromonas gingivalis fimbriae in relation to microbial pathogenesis. J Periodontal Res. 2004;39:136.
11 Amano A, Sharma A, Lee JY, et al. Structural domains of Porphyromonas gingivalis recombinant fimbrillin that mediate binding to salivary proline-rich protein and statherin. Infect Immun. 1996;64:1631.
12 American Academy of Periodontology: Proceedings of the 1996 World Workshop in Periodontics, 926, 1996.
13 Apse P, Ellen RP, Overall CM, et al. Microbiota and crevicular fluid collagenase activity in the osseointegrated dental implant sulcus: a comparison of sites in edentulous and partially edentulous patients. J Periodontal Res. 1989;24:96.
14 Arai M, Hamada N, Umemoto T. Purification and characterization of a novel secondary fimbrial protein from Porphyromonas gingivalis strain 381. FEMS Microbiol Lett. 2000;193:75.
15 Armitage GC. Development of a classification system for periodontal diseases and conditions. Ann Periodontol. 1999;4:1.
16 Arnim SS. The use of disclosing agents for measuring tooth cleanliness. J Periodontol. 1963;34:217.
17 Asakawa R, Komatsuzawa H, Kawai T, et al. Outer membrane protein 100, a versatile virulence factor of Actinobacillus actinomycetemcomitans. Mol Microbiol. 2003;50:1125.
18 Ashby MJ, Neale JE, Knott SJ, et al. Effect of antibiotics on non-growing planktonic cells and biofilms of Escherichia coli. J Antimicrob Chemother. 1994;33:443.
19 Asikainen S, Chen C, Slots J. Likelihood of transmitting Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in families with periodontitis. Oral Microbiol Immunol. 1996;11:387.
20 Bagaitkar J, Williams LR, Renaud DE, et al. Tobacco-induced alterations to Porphyromonas gingivalis-host interactions. Environ Microbiol. 2009;11:1242.
21 Barbour SE, Nakashima K, Zhang JB, et al. Tobacco and smoking: environmental factors that modify the host response (immune system) and have an impact on periodontal health. Crit Rev Oral Biol Med. 1997;8:437.
22 Beck JD, Koch GG, Zambon JJ, et al. Evaluation of oral bacteria as risk indicators for periodontitis in older adults. J Periodontol. 1992;63:93.
23 Beertsen W, de Haar SF, Mir M, et al. Gene symbol: CTSC. Disease: Papillon-Lefevre syndrome. Hum Genet. 2005;116:545.
24 Belay N, Johnson R, Rajagopal BS, et al. Methanogenic bacteria from human dental plaque. Appl Environ Microbiol. 1988;54:600.
25 Bergquist R. Parasitic infections affecting the oral cavity. Periodontol 2000. 2009;49:96.
26 Bollen CM, Mongardini C, Papaioannou W, et al. The effect of a one-stage full-mouth disinfection on different intra-oral niches. Clinical and microbiological observations. J Clin Periodontol. 1998;25:56.
27 Bollen CM, Papaioanno W, Van Eldere J, et al. The influence of abutment surface roughness on plaque accumulation and peri-implant mucositis. Clin Oral Implants Res. 1996;7:201.
28 Bostrom L, Bergstrom J, Dahlen G, et al. Smoking and subgingival microflora in periodontal disease. J Clin Periodontol. 2001;28:212.
29 Bowen WH. Nature of plaque. Oral Sci Rev. 1976;9:3.
30 Bower RC, Radny NR, Wall CD, et al. Clinical and microscopic findings in edentulous patients 3 years after incorporation of osseointegrated implant-supported bridgework. J Clin Periodontol. 1989;16:580.
31 Bradshaw DJ, Marsh PD, Watson GK, et al. Role of Fusobacterium nucleatum and coaggregation in anaerobe survival in planktonic and biofilm oral microbial communities during aeration. Infect Immun. 1998;66:4729.
32 Brailsford SR, Sheehy EC, Gilbert SC, et al. The microflora of the erupting first permanent molar. Caries Res. 2005;39:78.
33 Bramanti TE, Holt SC. Roles of porphyrins and host iron transport proteins in regulation of growth of Porphyromonas gingivalis W50. Journal of Bacteriology. 1991;173:7330.
34 Brogden KA, Guthmiller JM. Polymicrobial diseases involving viruses and bacteria. In: Brogden KA, Guthmiller JM, editors. Polymicrobial diseases. Washington DC: American Society Microbiology; 2002:201.
35 Brook I. Bacterial interference. Crit Rev Microbiol. 1999;25:155.
36 Brook I, Foote PA, Slots J. Immune response to Fusobacterium nucleatum, Prevotella intermedia and other anaerobes in children with acute tonsillitis. J Antimicrob Chemother. 1997;39:763.
37 Brooun A, Liu S, Lewis K. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2000;44:640.
38 Brown LJ, Loe H. Prevalence, extent, severity and progression of periodontal disease. Periodontol 2000. 1993;2:57.
39 Brown MR, Collier PJ, Gilbert P. Influence of growth rate on susceptibility to antimicrobial agents: modification of the cell envelope and batch and continuous culture studies. Antimicrob Agents Chemother. 1990;34:1623.
40 Brown MR, Williams P. The influence of environment on envelope properties affecting survival of bacteria in infections. Annu Rev Microbiol. 1985;39:527.
41 Brown SA, Whiteley M. A novel exclusion mechanism for carbon resource partitioning in Aggregatibacter actinomycetemcomitans. J Bacteriol. 2007;189:6407.
42 Brunette DM, Chehroudi B, Gould TRL. Electron microscopic observations on the effects of surface topography on the behaviour of cells attached to percutaneous and subcutaneous implants. In: Laney WR, Tonato M, editors. Tissue integration in Oral, Orthopedic and maxillofacial reconstruction. Chicago: Quintessence Books; 1982:21.
43 Busscher HJ, Norde W, van der Mei HC. Specific molecular recognition and nonspecific contributions to bacterial interaction forces. Appl Environ Microbiol. 2008;74:2559.
44 Busscher HJ, Sjollema J, van der Mei HC. Relative importance of surface free energy as a measure of hydrophobicity in bacterial adhesion to solid surfaces. In: Doyle RJ, Rosenberg M, editors. Microbial cell surface hydrophobicity. Washington DC: American Society for Microbiology; 1990:335.
45 Caimano MJ, Bourell KW, Bannister TD, et al. The Treponema denticola major sheath protein is predominantly periplasmic and has only limited surface exposure. Infect Immun. 1999;67:4072.
46 Cappuyns I, Gugerli P, Mombelli A. Viruses in periodontal disease—a review. Oral Dis. 2005;11:219.
47 Carlsson J. Symbiosis between host and microorganisms in the oral cavity. Scand J Infect Dis Suppl. 1980;Suppl 24:74.
48 Carlsson J. Microbiology of plaque associated periodontal disease. In: Lindhe J, editor. Textbook of clinical periodontology. Munksgaard: Munksgaard International Publishers, 1983.
49 Carlsson J, Grahnen H, Jonsson G. Lactobacilli and streptococci in the mouth of children. Caries Res. 1975;9:333.
50 Carlsson J, Grahnen H, Jonsson G, et al. Establishment of Streptococcus sanguis in the mouths of infants. Arch Oral Biol. 1970;15:1143.
51 Carlsson J, Soderholm G, Almfeldt I. Prevalence of Streptococcus sanguis and Streptococcus mutans in the mouth of persons wearing full-dentures. Arch Oral Biol. 1969;14:243.
52 Carranza FAJr, Saglie R, Newman MG, et al. Scanning and transmission electron microscopic study of tissue-invading microorganisms in localized juvenile periodontitis. J Periodontol. 1983;54:598.
53 Caufield PW, Dasanayake AP, Li Y, et al. Natural history of Streptococcus sanguinis in the oral cavity of infants: evidence for a discrete window of infectivity. Infect Immun. 2000;68:4018.
54 Caufield PW, Gibbons RJ. Suppression of Streptococcus mutans in the mouths of humans by a dental prophylaxis and topically-applied iodine. J Dent Res. 1979;58:1317.
55 Chen C, Slots J. Clonal analysis of Porphyromonas gingivalis by the arbitrarily primed polymerase chain reaction. Oral Microbiol Immunol. 1994;9:99.
56 Christersson LA, Albini B, Zambon JJ, et al. Tissue localization of Actinobacillus actinomycetemcomitans in human periodontitis. I. Light, immunofluorescence and electron microscopic studies. J Periodontol. 1987;58:529.
57 Christersson LA, Slots J, Zambon JJ, et al. Transmission and colonization of Actinobacillus actinomycetemcomitans in localized juvenile periodontitis patients. J Periodontol. 1985;56:127.
58 Christersson LA, Wikesjo UM, Albini B, et al. Tissue localization of Actinobacillus actinomycetemcomitans in human periodontitis. II. Correlation between immunofluorescence and culture techniques. J Periodontol. 1987;58:540.
59 Christersson LA, Zambon JJ, Genco RJ. Dental bacterial plaques. Nature and role in periodontal disease. J Clin Periodontol. 1991;18:441.
60 Cisar JO. Coaggregation reactions between oral bacteria: Studies of specific cell-to-cell adherence mediated by microbial lectins. In: Genco RJ, Mergenhagen S, editors. Host parasite interactions in periodontal diseases. Washington DC: American Society for Microbiology; 1982:121.
61 Contreras A, Slots J. Active cytomegalovirus infection in human periodontitis. Oral Microbiol Immunol. 1998;13:225.
62 Contreras A, Slots J. Herpesviruses in human periodontal disease. J Periodontal Res. 2000;35:3.
63 Contreras A, Umeda M, Chen C, et al. Relationship between herpesviruses and adult periodontitis and periodontopathic bacteria. J Periodontol. 1999;70:478.
64 Costerton JW, Lewandowski Z, Caldwell DE, et al. Microbial biofilms. Annu Rev Microbiol. 1995;49:711.
65 Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318.
66 Croen KD. Latency of the human herpesviruses. Annual Reviews of Medicine. 1991;42:61.
67 Cugini MA, Haffajee AD, Smith C, et al. The effect of scaling and root planing on the clinical and microbiological parameters of periodontal diseases: 12-month results. J Clin Periodontol. 2000;27:30.
68 Da Silva AM, Newman HN, Oakley DA. Psychosocial factors in inflammatory periodontal diseases. A review. J Clin Periodontol. 1995;22:516.
69 Danino J, Joachims HZ, Barak M. Predictive value of an adherence test for acute otitis media. Otolaryngol Head Neck Surg. 1998;118:400.
70 Danser MM, Timmerman MF, van Winkelhoff AJ, et al. The effect of periodontal treatment on periodontal bacteria on the oral mucous membranes. J Periodontol. 1996;67:478.
71 Danser MM, van Winkelhoff AJ, de Graaff J, et al. Short-term effect of full-mouth extraction on periodontal pathogens colonizing the oral mucous membranes. J Clin Periodontol. 1994;21:484.
72 Darveau RP, Tanner A, Page RC. The microbial challenge in periodontitis. Periodontol 2000. 1997;14:12.
73 De Soete M, De Keyser C, Pauwels M, et al. Outgrowth of cariogenic bacteria after initial periodontal therapy. J Dent Res. 2005. submitted
74 Diaz PI, Chalmers NI, Rickard AH, et al. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl Environ Microbiol. 2006;72:2837.
75 Diaz PI, Zilm PS, Rogers AH. The response to oxidative stress of Fusobacterium nucleatum grown in continuous culture. FEMS Microbiol Lett. 2000;187:31.
76 Dibart S, Skobe Z, Snapp KR, et al. Identification of bacterial species on or in crevicular epithelial cells from healthy and periodontally diseased patients using DNA-DNA hybridization. Oral Microbiol Immunol. 1998;13:30.
77 Dige I, Raarup MK, Nyengaard JR, et al. Actinomyces naeslundii in initial dental biofilm formation. Microbiology. 2009.
78 Doyle RJ, Rosenberg M, Drake D. Hydrophobicity of oral bacteria. In: Doyle RJ, Rosenberg M, editors. Microbial cell surface hydrophobicity. Washington DC: American Society for Microbiology, 1990.
79 Dzink JL, Gibbons RJ, Childs WCIII, et al. The predominant cultivable microbiota of crevicular epithelial cells. Oral Microbiol Immunol. 1989;4:1.
80 Dzink JL, Socransky SS, Haffajee AD. The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J Clin Periodontol. 1988;15:316.
81 Dzink JL, Tanner AC, Haffajee AD, et al. Gram negative species associated with active destructive periodontal lesions. J Clin Periodontol. 1985;12:648.
82 Edman DC, Keene HJ, Shklair IL, et al. Dental floss for implantation and sampling of Streptococcus mutans from approximal surfaces of human teeth. Arch Oral Biol. 1975;20:145.
83 Edwards AM, Jenkinson HF, Woodward MJ, et al. Binding properties and adhesion-mediating regions of the major sheath protein of Treponema denticola ATCC 35405. Infect Immun. 2005;73:2891.
84 Egland PG, Palmer RJJr, Kolenbrander PE. Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc Natl Acad Sci U S A. 2004;101:16917.
85 El Ahmer OR, Raza MW, Ogilvie MM, et al. Binding of bacteria to HEp-2 cells infected with influenza A virus. FEMS Immunol Med Microbiol. 1999;23:331.
86 Elangovan S, Margolis HC, Oppenheim FG, et al. Conformational changes in salivary proline-rich protein 1 upon adsorption to calcium phosphate crystals. Langmuir. 2007;23:11200.
87 Enersen M, Olsen I, Caugant DA. Genetic diversity of Porphyromonas gingivalis isolates recovered from single “refractory” periodontitis sites. Appl Environ Microbiol. 2008;74:5817.
88 Enersen M, Olsen I, Kvalheim O, et al. fimA genotypes and multilocus sequence types of Porphyromonas gingivalis from patients with periodontitis. J Clin Microbiol. 2008;46:31.
89 Evaldson G, Heimdahl A, Kager L, et al. The normal human anaerobic microflora. Scand J Infect Dis Suppl. 1982;35:9.
90 Evans P, Der G, Ford G, et al. Social class, sex, and age differences in mucosal immunity in a large community sample. Brain Behav Immun. 2000;14:41.
91 Fainstein V, Musher DM. Bacterial adherence to pharyngeal cells in smokers, nonsmokers, and chronic bronchitics. Infect Immun. 1979;26:178.
92 Faveri M, Figueiredo LC, Duarte PM, et al. Microbiological profile of untreated subjects with localized aggressive periodontitis. J Clin Periodontol. 2009;36:739.
93 Fenno JC, Muller KH, McBride BC. Sequence analysis, expression, and binding activity of recombinant major outer sheath protein (Msp) of Treponema denticola. J Bacteriol. 1996;178:2489.
94 Fine DH, Furgang D, Kaplan J, et al. Tenacious adhesion of Actinobacillus actinomycetemcomitans strain CU1000 to salivary-coated hydroxyapatite. Arch Oral Biol. 1999;44:1063.
95 Fletcher M. The physiological activity of bacteria attached to solid surfaces. Adv Microb Physiol. 1991;32:53.
96 Frank RM. Bacterial penetration in the apical pocket wall of advanced human periodontitis. J Periodontal Res. 1980;15:563.
97 Fransson C, Berglundh T, Lindhe J. The effect of age on the development of gingivitis. Clinical, microbiological and histological findings. J Clin Periodontol. 1996;23:379.
98 Frezzini C, Leao JC, Porter S. Cathepsin C involvement in the aetiology of Papillon-Lefevre syndrome. Int J Paediatr Dent. 2004;14:466.
99 Fujiwara T, Sasada E, Mima N, et al. Caries prevalence and salivary mutans streptococci in 0-2-year-old children of Japan. Community Dent Oral Epidemiol. 1991;19:151.
100 Fullmer SC, Preshaw PM, Heasman PA, et al. Smoking cessation alters subgingival microbial recolonization. J Dent Res. 2009;88:524.
101 Furst MM, Salvi GE, Lang NP, et al. Bacterial colonization immediately after installation on oral titanium implants. Clin Oral Implants Res. 2007;18:501.
102 Furuichi Y, Lindhe J, Ramberg P, et al. Patterns of de novo plaque formation in the human dentition. J Clin Periodontol. 1992;19:423.
103 Genco CA, Arko RJ. Animal chamber models for study of host-parasite interactions. Methods Enzymol. 1994;235:120.
104 Genco RJ, Ho AW, Grossi SG, et al. Relationship of stress, distress and inadequate coping behaviors to periodontal disease. J Periodontol. 1999;70:711.
105 Gibson GR, Roberfroid MB. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr. 1995;125:1401.
106 Gilbert P, Allison DG. Biofilms and their resistance towards antimicrobial agents. In: Newman HN, Wilson M, editors. Dental plaque revisited. Cambridge: Cambridge University Press; 1999:125.
107 Gilthorpe MS, Zamzuri AT, Griffiths GS, et al. Unification of the “burst” and “linear” theories of periodontal disease progression: a multilevel manifestation of the same phenomenon. J Dent Res. 2003;82:200.
108 Giuliana G, Pizzo G, Milici ME, et al. In vitro antifungal properties of mouthrinses containing antimicrobial agents. J Periodontol. 1997;68:729.
109 Gmür R, Lüthi-Schaller H. A combined immunofluorescence and fluorescent in situ hybridization assay for single cell analyses of dental plaque microorganisms. J Microbiol Methods. 2007;69:402.
110 Goobes G, Goobes R, Schueler-Furman O, et al. Folding of the C-terminal bacterial binding domain in statherin upon adsorption onto hydroxyapatite crystals. Proc Natl Acad Sci U S A. 2006;103:16083.
111 Goodson JM, Tanner A, McArdle S, et al. Multicenter evaluation of tetracycline fiber therapy. III. Microbiological response. J Periodontal Res. 1991;26:440.
112 Goodson JM, Tanner AC, Haffajee AD, et al. Patterns of progression and regression of advanced destructive periodontal disease. J Clin Periodontol. 1982;9:472.
113 Grenier D. Nutritional interactions between two suspected periodontopathogens, Treponema denticola and Porphyromonas gingivalis. Infect Immun. 1992;60:5298.
114 Grenier D, Mayrand D. [Studies of mixed anaerobic infections involving Bacteroides gingivalis]. Can J Microbiol. 1983;29:612.
115 Griffen AL, Leys EJ, Fuerst PA. Strain identification of Actinobacillus actinomycetemcomitans using the polymerase chain reaction. Oral Microbiol Immunol. 1992;7:240.
116 Grossi SG, Genco RJ, Machtei EE, et al. Assessment of risk for periodontal disease. II. Risk indicators for alveolar bone loss. J Periodontol. 1995;66:23.
117 Grossi SG, Zambon JJ, Ho AW, et al. Assessment of risk for periodontal disease. I. Risk indicators for attachment loss. J Periodontol. 1994;65:260.
118 Guy SC, McQuade MJ, Scheidt MJ, et al. In vitro attachment of human gingival fibroblasts to endosseous implant materials. J Periodontol. 1993;64:542.
119 Haapasalo M, Muller KH, Uitto VJ, et al. Characterization, cloning, and binding properties of the major 53-kilodalton Treponema denticola surface antigen. Infect Immun. 1992;60:2058.
120 Haffajee AD, Cugini MA, Dibart S, et al. Clinical and microbiological features of subjects with adult periodontitis who responded poorly to scaling and root planing. J Clin Periodontol. 1997;24:767.
121 Haffajee AD, Cugini MA, Dibart S, et al. The effect of SRP on the clinical and microbiological parameters of periodontal diseases. J Clin Periodontol. 1997;24:324.
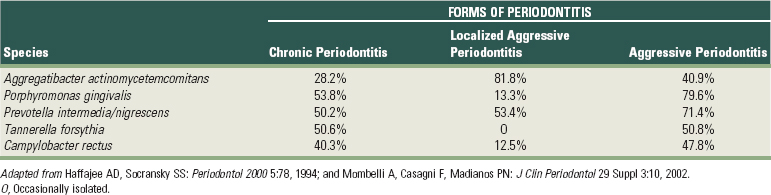
122 Haffajee AD, Socransky SS. Microbial etiological agents of destructive periodontal diseases. Periodontol 2000. 1994;5:78.
123 Haffajee AD, Socransky SS. Relationship of cigarette smoking to the subgingival microbiota. J Clin Periodontol. 2001;28:377.
124 Hafstrom CA, Wikstrom MB, Renvert SN, et al. Effect of treatment on some periodontopathogens and their antibody levels in periodontal abscesses. J Periodontol. 1994;65:1022.
125 Hakansson A, Kidd A, Wadell G, et al. Adenovirus infection enhances in vitro adherence of Streptococcus pneumoniae. Infect Immun. 1994;62:2707.
126 Hamada N, Sojar HT, Cho MI, et al. Isolation and characterization of a minor fimbria from Porphyromonas gingivalis. Infect Immun. 1996;64:4788.
127 Hamada S, Amano A, Kimura S, et al. The importance of fimbriae in the virulence and ecology of some oral bacteria. Oral Microbiol Immunol. 1998;13:129.
128 Hammarstedt L, Lindquist D, Dahlstrand H, et al. Human papillomavirus as a risk factor for the increase in incidence of tonsillar cancer. Int J Cancer. 2006;119:2620.
129 Hannig C, Attin T, Hannig M, et al. Immobilisation and activity of human alpha-amylase in the acquired enamel pellicle. Arch Oral Biol. 2004;49:469.
130 Hannig C, Hannig M, Rehmer O, et al. Fluorescence microscopic visualization and quantification of initial bacterial colonization on enamel in situ. Arch Oral Biol. 2007;52:1048.
131 Hannig C, Hoch J, Becker K, et al. Lysozyme activity in the initially formed in situ pellicle. Arch Oral Biol. 2005;50:821.
132 Hannig C, Spitzmuller B, Hannig M. Characterisation of lysozyme activity in the in situ pellicle using a fluorimetric assay. Clin Oral Investig. 2009;13:15.
133 Hannig C, Spitzmuller B, Knausenberger S, et al. Detection and activity of peroxidase in the in situ formed enamel pellicle. Arch Oral Biol. 2008;53:849.
134 Hannig M. Ultrastructural investigation of pellicle morphogenesis at two different intraoral sites during a 24-h period. Clin Oral Investig. 1999;3:88.
135 Hannig M, Khanafer AK, Hoth-Hannig W, et al. Transmission electron microscopy comparison of methods for collecting in situ formed enamel pellicle. Clin Oral Investig. 2005;9:30.
136 Harper DS, Robinson PJ. Correlation of histometric, microbial, and clinical indicators of periodontal disease status before and after root planing. J Clin Periodontol. 1987;14:190.
137 Hart TC, Marazita ML, Schenkein HA, et al. Re-interpretation of the evidence for X-linked dominant inheritance of juvenile periodontitis. J Periodontol. 1992;63:169.
138 Haubek D, Poulsen K, Kilian M. Microevolution and patterns of dissemination of the JP2 clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans. Infect Immun. 2007;75:3080.
139 Helmerhorst EJ, Hodgson R, van’t HW, et al. The effects of histatin-derived basic antimicrobial peptides on oral biofilms. J Dent Res. 1999;78:1245.
140 Hermansson M. The DLVO theory in microbial adhesion. Colloids and Surfaces B-Biointerfaces. 1999;14:105.
141 Herrera D, Roldan S, Gonzalez I, et al. The periodontal abscess (I). Clinical and microbiological findings. J Clin Periodontol. 2000;27:387.
142 Herrera D, Roldan S, Sanz M. The periodontal abscess: a review. J Clin Periodontol. 2000;27:377.
143 Hewitt C, McCormick D, Linden G, et al. The role of cathepsin C in Papillon-Lefevre syndrome, prepubertal periodontitis, and aggressive periodontitis. Hum Mutat. 2004;23:222.
144 Hillman JD, Socransky SS, Shivers M. The relationships between streptococcal species and periodontopathic bacteria in human dental plaque. Arch Oral Biol. 1985;30:791.
145 Hobbs CG, Sterne JA, Bailey M, et al. Human papillomavirus and head and neck cancer: a systematic review and meta-analysis. Clin Otolaryngol. 2006;31:259.
146 Hoepelman AI, Tuomanen EI. Consequences of microbial attachment: directing host cell functions with adhesins. Infect Immun. 1992;60:1729.
147 Holm-Pedersen P, Agerbaek N, Theilade E. Experimental gingivitis in young and elderly individuals. J Clin Periodontol. 1975;2:14.
148 Hsu SD, Cisar JO, Sandberg AL, et al. Adhesive properties of viridans streptococcal species. Microb Ecol Health Dis. 1994:125.
149 Hultin M, Gustafsson A, Hallstrom H, et al. Microbiological findings and host response in patients with peri-implantitis. Clin Oral Implants Res. 2002;13:349.
150 Hultin M, Gustafsson A, Klinge B. Long-term evaluation of osseointegrated dental implants in the treatment of partly edentulous patients. J Clin Periodontol. 2000;27:128.
151 Hyyppä T, Paunio K. The plaque-inhibiting effects of copper amalgam. J Clin Periodontol. 1977;4:231.
152 Imamura T, Potempa J, Travis J. Comparison of pathogenic properties between two types of arginine-specific cysteine proteinases (gingipains-R) from Porphyromonas gingivalis. Microb Pathog. 2000;29:155.
153 Isogai E, Isogai H, Sawada H, et al. Bacterial adherence to gingival epithelial cells of rats with naturally occurring gingivitis. J Periodontol. 1986;57:225.
154 Jacobson SE, Crawford JJ, McFall WRJr. Oral physiotherapy of the tongue and palate: relationship to plaque control. J Am Dent Assoc. 1973;87:134.
155 Jakubovics NS, Gill SR, Iobst SE, et al. Regulation of gene expression in a mixed-genus community: stabilized arginine biosynthesis in Streptococcus gordonii by coaggregation with Actinomyces naeslundii. J Bacteriol. 2008;190:3646.
156 Jakubovics NS, Stromberg N, van Dolleweerd CJ, et al. Differential binding specificities of oral streptococcal antigen I/II family adhesins for human or bacterial ligands. Mol Microbiol. 2005;55:1591.
157 Jenkinson HF, Demuth DR. Structure, function and immunogenicity of streptococcal antigen I/II polypeptides. Mol Microbiol. 1997;23:183.
158 Jette AM, Feldman HA, Tennstedt SL. Tobacco use: a modifiable risk factor for dental disease among the elderly. Am J Public Health. 1993;83:1271.
159 Johnson GK, Hill M. Cigarette smoking and the periodontal patient. J Periodontol. 2004;75:196.
160 Johnson JD, Chen R, Lenton PA, et al. Persistence of extracrevicular bacterial reservoirs after treatment of aggressive periodontitis. J Periodontol. 2008;79:2305.
161 Kaldahl WB, Johnson GK, Patil KD, et al. Levels of cigarette consumption and response to periodontal therapy. J Periodontol. 1996;67:675.
162 Kaplan CW, Lux R, Haake SK, et al. The Fusobacterium nucleatum outer membrane protein RadD is an arginine-inhibitable adhesin required for inter-species adherence and the structured architecture of multispecies biofilm. Mol Microbiol. 2009;71:35.
163 Kaplan JB, Ragunath C, Ramasubbu N, et al. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous beta-hexosaminidase activity. J Bacteriol. 2003;185:4693.
164 Kaufman J, DiRienzo JM. Isolation of a corncob (coaggregation) receptor polypeptide from Fusobacterium nucleatum. Infect Immun. 1989;57:331.
165 Keijser BJ, Zaura E, Huse SM, et al. Pyrosequencing analysis of the oral microflora of healthy adults. J Dent Res. 2008;87:1016.
166 Kinder SA, Holt SC. Characterization of coaggregation between Bacteroides gingivalis T22 and Fusobacterium nucleatum T18. Infect Immun. 1989;57:3425.
167 Kohavi D, Greenberg RS, Raviv E, et al. Subgingival and supragingival microbial flora around healthy osseointegrated implants in partially edentulous patients. Int J Oral Maxillofac Implants. 1994:673.
168 Kohler B, Krasse B, Carlen A. Adherence and Streptococcus mutans infections: in vitro study with saliva from noninfected and infected preschool children. Infect Immun. 1981;34:633.
169 Koka S, Razzoog ME, Bloem TJ, et al. Microbial colonization of dental implants in partially edentulous subjects. J Prosthet Dent. 1993;70:141.
170 Kolenbrander PE. Surface recognition among oral bacteria: multigeneric coaggregations and their mediators. Crit Rev Microbiol. 1989;17:137.
171 Kolenbrander PE, Andersen RN. Inhibition of coaggregation between Fusobacterium nucleatum and Porphyromonas (Bacteroides) gingivalis by lactose and related sugars. Infect Immun. 1989;57:3204.
172 Kolenbrander PE, Andersen RN, Moore LV. Intrageneric coaggregation among strains of human oral bacteria: potential role in primary colonization of the tooth surface. Appl Environ Microbiol. 1990;56:3890.
173 Kolenbrander PE, Ganeshkumar N, Cassels FJ, et al. Coaggregation: specific adherence among human oral plaque bacteria. FASEB J. 1993;7:406.
174 Kolenbrander PE, London J. Adhere today, here tomorrow: oral bacterial adherence. Journal of Bacteriology. 1993;175:3247.
175 Kolenbrander PE, Palmer RJJr, Rickard AH, et al. Bacterial interactions and successions during plaque development. Periodontol 2000. 2006;42:47.
176 Kolenbrander PE, Parrish KD, Andersen RN, et al. Intergeneric coaggregation of oral Treponema spp. with Fusobacterium spp. and intrageneric coaggregation among Fusobacterium spp. Infect Immun. 1995;63:4584.
177 Kononen E. Oral colonization by anaerobic bacteria during childhood: role in health and disease. Oral Dis. 1999;5:278.
178 Kononen E. Development of oral bacterial flora in young children. Ann Med. 2000;32:107.
179 Kononen E, Asikainen S, Jousimies-Somer H. The early colonization of gram-negative anaerobic bacteria in edentulous infants. Oral Microbiol Immunol. 1992;7:28.
180 Kornman KS, Loe H. The role of local factors in the etiology of periodontal diseases. Periodontol 2000. 1993;2:83.
181 Kornman KS, Loesche WJ. Effects of estradiol and progesterone on Bacteroides melaninogenicus and Bacteroides gingivalis. Infect Immun. 1982;35:256.
182 Kornman KS, Robertson PB. Clinical and microbiological evaluation of therapy for juvenile periodontitis. J Periodontol. 1985;56:443.
183 Kremer BH, Loos BG, van d V, et al. Peptostreptococcus micros smooth and rough genotypes in periodontitis and gingivitis. J Periodontol. 2000;71:209.
184 Kreth J, Zhang Y, Herzberg MC. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J Bacteriol. 2008;190:4632.
185 Kroes I, Lepp PW, Relman DA. Bacterial diversity within the human subgingival crevice. Proc Natl Acad Sci U S A. 1999;96:14547.
186 Kuboniwa M, Hendrickson EL, Xia Q, et al. Proteomics of Porphyromonas gingivalis within a model oral microbial community. BMC Microbiol. 2009;9:98.
187 Kumamoto CA, Vinces MD. Alternative Candida albicans lifestyles: growth on surfaces. Annu Rev Microbiol. 2005;59:113.
188 Kuritzkes DR, Walker BD. HIV-1 pathogenesis, clinical manifestations, and treatment. In: Knipe DM, Howley PM, editors. Fields virology. Philadelphia: Lippincott, Williams and Wilkins; 2007:2187.
189 Lai CH, Listgarten MA, Shirakawa M, et al. Bacteroides forsythus in adult gingivitis and periodontitis. Oral Microbiol Immunol. 1987;2:152.
190 Laine ML, Appelmelk BJ, van Winkelhoff AJ. Novel polysaccharide capsular serotypes in Porphyromonas gingivalis. J Periodontal Res. 1996;31:278.
191 Laine ML, Appelmelk BJ, van Winkelhoff AJ. Prevalence and distribution of six capsular serotypes of Porphyromonas gingivalis in periodontitis patients. J Dent Res. 1997;76:1840.
192 Laine ML, van Winkelhoff AJ. Virulence of six capsular serotypes of Porphyromonas gingivalis in a mouse model. Oral Microbiol Immunol. 1998;13:322.
193 Lamont RJ, Chan A, Belton CM, et al. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun. 1995;63:3878.
194 Lang NP, Cumming BR, Loe H. Toothbrushing frequency as it relates to plaque development and gingival health. J Periodontol. 1973;44:396.
195 Lappin-Scott HM, Claude E. Zobell—his life and contributions to biofilm microbiology. In: Bell CR, Brylinsky M, Johnson-Green P, editors. Microbial biosystems: new frontiers. Halifax, Canada: Atlantic Canada Society for Microbial Ecology; 1999:45.
196 Law V, Seow WK. A longitudinal controlled study of factors associated with mutans streptococci infection and caries lesion initiation in children 21 to 72 months old. Pediatr Dent. 2006;28:58.
197 Law V, Seow WK, Townsend G. Factors influencing oral colonization of mutans streptococci in young children. Aust Dent J. 2007;52:93.
198 Lekholm U, Ericsson I, Adell R, et al. The condition of the soft tissues at tooth and fixture abutments supporting fixed bridges. A microbiological and histological study. J Clin Periodontol. 1986;13:558.
199 Leknes KN, Lie T, Selvig KA. Root grooves: a risk factor in periodontal attachment loss. J Periodontol. 1994;65:859.
200 Leknes KN, Lie T, Wikesjo UM, et al. Influence of tooth instrumentation roughness on gingival tissue reactions. J Periodontol. 1996;67:197.
201 Leknes KN, Lie T, Wikesjo UM, et al. Influence of tooth instrumentation roughness on subgingival microbial colonization. J Periodontol. 1994;65:303.
202 Leonhardt A, Renvert S, Dahlen G. Microbial findings at failing implants. Clin Oral Implants Res. 1999;10:339.
203 Lepp PW, Brinig MM, Ouverney CC, et al. Methanogenic Archaea and human periodontal disease. Proc Natl Acad Sci U S A. 2004;101:6176.
204 Li L, Matevski D, Aspiras M, et al. Two epithelial cell invasion-related loci of the oral pathogen Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol. 2004;19:16.
205 Li L, Redding S, Dongari-Bagtzoglou A. Candida glabrata: an emerging oral opportunistic pathogen. J Dent Res. 2007;86:204.
206 Liljemark WF, Bloomquist CG, Germaine GR. Effect of bacterial aggregation on the adherence of oral streptococci to hydroxyapatite. Infect Immun. 1981;31:935.
207 Lin X, Lamont RJ, Wu J, et al. Role of differential expression of streptococcal arginine deiminase in inhibition of fimA expression in Porphyromonas gingivalis. J Bacteriol. 2008;190:4367.
208 Lindquist LW, Carlsson GE, Jemt T. Association between marginal bone loss around osseointegrated mandibular implants and smoking habits: a 10-year follow-up study. J Dent Res. 1997;76:1667.
209 Listgarten MA. Structure of the microbial flora associated with periodontal health and disease in man. A light and electron microscopic study. J Periodontol. 1976;47:1.
210 Listgarten MA, Hellden L. Relative distribution of bacteria at clinically healthy and periodontally diseased sites in humans. J Clin Periodontol. 1978;5:115.
211 Listgarten MA, Socransky SS. Ultrastructural characteristics of a spirochete in the lesion of acute necrotizing ulcerative gingivostomatitis (Vincent’s infection). Arch Oral Biol. 1964;16:95.
212 Listgarten MA, Socransky SS. Electron microscopy as an aid in the taxonomic differentiation of oral spirochetes. Arch Oral Biol. 1965;10:127.
213 Loe H. Periodontal diseases: a brief historical perspective. Periodontol 2000. 1993;2:7.
214 Loë H, Anerud A, Boysen H, et al. The natural history of periodontal disease in man. The rate of periodontal destruction before 40 years of age. J Periodontol. 1978;49:607.
215 Loë H, Brown LJ. Early onset periodontitis in the United States of America. J Periodontol. 1991;62:608.
216 Loe H, Theilade E, Jensen SB. Experimental gingivitis in man. J Periodontol. 1965;36:177.
217 Loesche WJ. Importance of nutrition in gingival crevice microbial ecology. Periodontics. 1968;6:245.
218 Loesche WJ. Chemotherapy of dental plaque infections. Oral Sci Rev. 1976;9:65.
219 Loesche WJ, Svanberg ML, Pape HR. Intraoral transmission of Streptococcus mutans by a dental explorer. J Dent Res. 1979;58:1765.
220 Loesche WJ, Syed SA, Schmidt E, et al. Bacterial profiles of subgingival plaques in periodontitis. J Periodontol. 1985;56:447.
221 Loimaranta V, Jakubovics NS, Hytonen J, et al. Fluid- or surface-phase human salivary scavenger protein gp340 exposes different bacterial recognition properties. Infect Immun. 2005;73:2245.
222 Loomer PM. Microbiological diagnostic testing in the treatment of periodontal diseases. Periodontol 2000. 2004;34:49.
223 Lovdal A, Arno A, Waerhaug J. Incidence of clinical manifestations of periodontal disease in light of oral hygiene and calculus formation. J Am Dent Assoc. 1958;56:21.
224 Lucht E, Evengard B, Skott J, et al. Entamoeba gingivalis in human immunodeficiency virus type 1-infected patients with periodontal disease. Clin Infect Dis. 1998;27:471.
225 Lundgren T, Renvert S. Periodontal treatment of patients with Papillon-Lefevre syndrome: a 3-year follow-up. J Clin Periodontol. 2004;31:933.
226 MacFarlane GD, Herzberg MC, Wolff LF, et al. Refractory periodontitis associated with abnormal polymorphonuclear leukocyte phagocytosis and cigarette smoking. J Periodontol. 1992;63:908.
227 MacFarlane TW, Samaranayake LP. Systemic infections. In: Jones JH, Manson DH, editors. Oral manifestations of systemic disease. London: Balliere Tindall; 1990:339.
228 Maeda K, Nagata H, Nonaka A, et al. Oral streptococcal glyceraldehyde-3-phosphate dehydrogenase mediates interaction with Porphyromonas gingivalis fimbriae. Microbes Infect. 2004;6:1163.
229 Maiden MF, Tanner A, McArdle S, et al. Tetracycline fiber therapy monitored by DNA probe and cultural methods. J Periodontal Res. 1991;26:452.
230 Mandlik A, Swierczynski A, Das A, et al. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 2008;16:33.
231 Manor A, Lebendiger M, Shiffer A, et al. Bacterial invasion of periodontal tissues in advanced periodontitis in humans. J Periodontol. 1984;55:567.
232 Marsh PD. Microbial ecology of dental plaque and its significance in health and disease. Adv Dent Res. 1994;8:263.
233 Mayrand D, McBride BC. Exological relationships of bacteria involved in a simple, mixed anaerobic infection. Infect Immun. 1980;27:44.
234 McBride BC, Van der Hoeven JS. Role of interbacterial adherence in colonization of the oral cavities of gnotobiotic rats infected with Streptococcus mutans and Veillonella alcalescens. Infect Immun. 1981;33:467.
235 Mengel R, Stelzel M, Hasse C, et al. Osseointegrated implants in patients treated for generalized severe adult periodontitis. An interim report. J Periodontol. 1996;67:782.
236 Meyer DH, Sreenivasan PK, Fives-Taylor PM. Evidence for invasion of a human oral cell line by Actinobacillus actinomycetemcomitans. Infect Immun. 1991;59:2719.
237 Michalowicz BS. Genetic and heritable risk factors in periodontal disease. J Periodontol. 1994;65:479.
238 Michalowicz BS, Ronderos M, Camara-Silva R, et al. Human herpesviruses and Porphyromonas gingivalis are associated with juvenile periodontitis. J Periodontol. 2000;71:981.
239 Mierau HD. [Relations between plaque formation, tooth surface roughness and self-cleaning]. Dtsch Zahnarztl Z. 1984;39:691.
240 Mierau HD, Singer D. [Reproducibility of plaque formation in the dento-gingival region (3)]. Dtsch Zahnarztl Z. 1978;33:566.
241 Milgrom P, Riedy CA, Weinstein P, et al. Dental caries and its relationship to bacterial infection, hypoplasia, diet, and oral hygiene in 6- to 36-month-old children. Community Dent Oral Epidemiol. 2000;28:295.
242 Miller PD. Root coverage with the free gingival graft. Factors associated with incomplete coverage. J Periodontol. 1987;58:674.
243 Mombelli A, Casagni F, Madianos PN. Can presence or absence of periodontal pathogens distinguish between subjects with chronic and aggressive periodontitis? A systematic review. J Clin Periodontol. 2002;29(Suppl 3):10.
244 Mombelli A, Lang NP, Burgin WB, et al. Microbial changes associated with the development of puberty gingivitis. J Periodontal Res. 1990;25:331.
245 Mombelli A, Lehmann B, Tonetti M, et al. Clinical response to local delivery of tetracycline in relation to overall and local periodontal conditions. J Clin Periodontol. 1997;24:470.
246 Mombelli A, Marxer M, Gaberthuel T, et al. The microbiota of osseointegrated implants in patients with a history of periodontal disease. J Clin Periodontol. 1995;22:124.
247 Moore WE. Microbiology of periodontal disease. J Periodontal Res. 1987;22:335.
248 Moore WE, Holdeman LV. Special problems associated with the isolation and identification of intestinal bacteria in fecal flora studies. Am J Clin Nutr. 1974;27:1450.
249 Moore WE, Holdeman LV, Cato EP, et al. Comparative bacteriology of juvenile periodontitis. Infect Immun. 1985;48:507.
250 Moore WE, Moore LV. The bacteria of periodontal diseases. Periodontol 2000. 1994;5:66.
251 Mouton C, Reynolds HS, Genco RJ. Characterization of tufted streptococci isolated from the “corn cob” configuration of human dental plaque. Infect Immun. 1980;27:235.
252 Nelson KE, Fleischmann RD, DeBoy RT, et al. Complete genome sequence of the oral pathogenic Bacterium porphyromonas gingivalis strain W83. J Bacteriol. 2003;185:5591.
253 Newman HN. The approximal apical border of plaque on children’s teeth. 1. Morphology, structure and cell content. J Periodontol. 1979;50:561.
254 Newman MG, Socransky SS. Predominant cultivable microbiota in periodontosis. J Periodontal Res. 1977;12:120.
255 Newman MG, Socransky SS, Savitt ED, et al. Studies of the microbiology of periodontosis. J Periodontol. 1976;47:373.
256 Nishiyama S, Murakami Y, Nagata H, et al. Involvement of minor components associated with the FimA fimbriae of Porphyromonas gingivalis in adhesive functions. Microbiology. 2007;153:1916.
257 Nowzari H, MacDonald ES, Flynn J, et al. The dynamics of microbial colonization of barrier membranes for guided tissue regeneration. J Periodontol. 1996;67:694.
258 Nyvad B, Fejerskov O. Scanning electron microscopy of early microbial colonization of human enamel and root surfaces in vivo. Scand J Dent Res. 1987;95:287.
259 Nyvad B, Kilian M. Microflora associated with experimental root surface caries in humans. Infect Immun. 1990;58:1628.
260 O’Brien-Simpson NM, Veith PD, Dashper SG, et al. Antigens of bacteria associated with periodontitis. Periodontol 2000. 2004;35:101.
261 Oberste MS, Maher K, Kilpatrick DR, et al. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J Virol. 1999;73:1941.
262 Ofek I, Doyle RJ, editors. Bacterial adhesion to cells and tissues. London: Chapman and Hall, 1994.
263 Ong ES, Newman HN, Wilson M, et al. The occurrence of periodontitis-related microorganisms in relation to titanium implants. J Periodontol. 1992;63:200.
264 Ouverney CC, Armitage GC, Relman DA. Single-cell enumeration of an uncultivated TM7 subgroup in the human subgingival crevice. Appl Environ Microbiol. 2003;69:6294.
265 Pallansch M, Roos R. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses. In: Knipe DM, Howley PM, editors. Fields virology. Philadelphia: Lippincott, Williams and Wilkins; 2007:840.
266 Palmer RJJr, Gordon SM, Cisar JO, et al. Coaggregation-mediated interactions of streptococci and actinomyces detected in initial human dental plaque. J Bacteriol. 2003;185:3400.
267 Papaioannou W, Quirynen M, van Steenberghe D. The influence of periodontitis on the subgingival flora around implants in partially edentulous patients. Clin Oral Implants Res. 1996;7:405.
268 Park Y, James CE, Yoshimura F, et al. Expression of the short fimbriae of Porphyromonas gingivalis is regulated in oral bacterial consortia. FEMS Microbiol Lett. 2006;262:65.
269 Park Y, Simionato MR, Sekiya K, et al. Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect Immun. 2005;73:3983.
270 Patton LL, Phelan JA, Ramos-Gomez FJ, et al. Prevalence and classification of HIV-associated oral lesions. Oral Dis. 2002;8(Suppl 2):98.
271 Pearce C, Bowden GH, Evans M, et al. Identification of pioneer viridans streptococci in the oral cavity of human neonates. Journal of Medical Microbiology. 1995;42:67.
272 Pearce MA, Devine DA, Dixon RA, et al. Genetic heterogeneity in Prevotella intermedia, Prevotella nigrescens, Prevotella corporis and related species isolated from oral and nonoral sites. Oral Microbiol Immunol. 2000;15:89.
273 Pellett PE, Roizman B. The family Herpesviridae: a brief introduction. In: Knipe DM, Howley PM, editors. Fields virology. Philadelphia: Lippincott, Williams and Wilkins; 2007:2479.
274 Perez BA, Planet PJ, Kachlany SC, et al. Genetic analysis of the requirement for flp-2, tadV, and rcpB in Actinobacillus actinomycetemcomitans biofilm formation. J Bacteriol. 2006;188:6361.
275 Perry JA, Jones MB, Peterson SN, et al. Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence. Mol Microbiol. 2009.
276 Petersilka GJ, Ehmke B, Flemmig TF. Antimicrobial effects of mechanical debridement. Periodontol 2000. 2002;28:56.
277 Pfaller MA. Nosocomial candidiasis: emerging species, reservoirs, and modes of transmission. Clin Infect Dis. 1996;22(Suppl 2):S89.
278 Planet PJ, Kachlany SC, Fine DH, et al. The widespread colonization island of Actinobacillus actinomycetemcomitans. Nat Genet. 2003;34:193.
279 Porter SR, Luker J, Scully C, et al. Orofacial manifestations of a group of British patients infected with HIV-1. J Oral Pathol Med. 1989;18:47.
280 Potempa J, Banbula A, Travis J. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontol 2000. 2000;24:153.
281 Potempa J, Pike R, Travis J. Titration and mapping of the active site of cysteine proteinases from Porphyromonas gingivalis (gingipains) using peptidyl chloromethanes. Biol Chem. 1997;378:223.
282 Potempa M, Potempa J, Kantyka T, et al. Interpain A, a cysteine proteinase from Prevotella intermedia, inhibits complement by degrading complement factor C3. PLoS Pathog. 2009;5:e1000316.
283 Prakobphol A, Xu F, Hoang VM, et al. Salivary agglutinin, which binds Streptococcus mutans and Helicobacter pylori, is the lung scavenger receptor cysteine-rich protein gp-340. J Biol Chem. 2000;275:39860.
284 Pratten J, Barnett P, Wilson M. Composition and susceptibility to chlorhexidine of multispecies biofilms of oral bacteria. Appl Environ Microbiol. 1998;64:3515.
285 Preber H, Bergstrom J, Linder LE. Occurrence of periopathogens in smoker and non-smoker patients. J Clin Periodontol. 1992;19:667.
286 Qandil R, Sandhu HS, Matthews DC. Tobacco smoking and periodontal diseases. J Can Dent Assoc. 1997;63:187.
287 Quinn SM, Zhang JB, Gunsolley JC, et al. Influence of smoking and race on immunoglobulin G subclass concentrations in early-onset periodontitis patients. Infect Immun. 1996;64:2500.
288 Quirynen M, Anatomical and inflammatory factors influence bacterial plaque growth and retention in man, 1986
289 Quirynen M, Bollen CM. The influence of surface roughness and surface-free energy on supra- and subgingival plaque formation in man. A review of the literature. J Clin Periodontol. 1995;22:1.
290 Quirynen M, Bollen CM. The influence of surface roughness and surface-free energy on supra- and subgingival plaque formation in man. A review of the literature. J Clin Periodontol. 1995;22:1.
291 Quirynen M, Bollen CM, Papaioannou W, et al. The influence of titanium abutment surface roughness on plaque accumulation and gingivitis: short-term observations. Int J Oral Maxillofac Implants. 1996;11:169.
292 Quirynen M, Bollen CM, Vandekerckhove BN, et al. Full- vs. partial-mouth disinfection in the treatment of periodontal infections: short-term clinical and microbiological observations. J Dent Res. 1995;74:1459.
293 Quirynen M, De Soete M, Dierickx K, et al. The intra-oral translocation of periodontopathogens jeopardises the outcome of periodontal therapy. A review of the literature. J Clin Periodontol. 2001;28:499.
294 Quirynen M, Dekeyser C, van Steenberghe D. The influence of gingival inflammation, tooth type, and timing on the rate of plaque formation. J Periodontol. 1991;62:219.
295 Quirynen M, Gijbels F, Jacobs R. An infected jawbone site compromising successful osseointegration. Periodontol 2000. 2003;33:129.
296 Quirynen M, Gizani S, Mongardini C, et al. The effect of periodontal therapy on the number of cariogenic bacteria in different intra-oral niches. J Clin Periodontol. 1999;26:322.
297 Quirynen M, Listgarten MA. Distribution of bacterial morphotypes around natural teeth and titanium implants ad modum Branemark. Clin Oral Implants Res. 1990;1:8.
298 Quirynen M, Marechal M, Busscher HJ, et al. The influence of surface free-energy on planimetric plaque growth in man. J Dent Res. 1989;68:796.
299 Quirynen M, Papaioannou W, van Steenbergen TJ, et al. Adhesion of Porphyromonas gingivalis strains to cultured epithelial cells from patients with a history of chronic adult periodontitis or from patients less susceptible to periodontitis. J Periodontol. 2001;72:626.
300 Quirynen M, Teughels W, De Soete M, et al. Topical antiseptics and antibiotics in the initial therapy of chronic adult periodontitis: microbiological aspects. Periodontol 2000. 2002;28:72.
301 Quirynen M, van der Mei HC, Bollen CM, et al. An in vivo study of the influence of the surface roughness of implants on the microbiology of supra- and subgingival plaque. J Dent Res. 1993;72:1304.
302 Quirynen M, van Steenberghe D. Is early plaque growth rate constant with time? J Clin Periodontol. 1989;16:278.
303 Quirynen M, Vogels R, Pauwels M, et al. Initial subgingival colonization of pristine pockets in an established environment. J Dent Res. 2005. submitted
304 Quirynen M, Vogels R, Peeters W, et al. Dynamics of initial subgingival colonization of peri-implant pockets. Clin Oral Implants Res. 2005. submitted
305 Quirynen M, Vogels R, Peeters W, et al. Dynamics of initial subgingival colonization of “pristine” peri-implant pockets. Clin Oral Implants Res. 2006;17:25.
306 Ramberg P, Axelsson P, Lindhe J. Plaque formation at healthy and inflamed gingival sites in young individuals. J Clin Periodontol. 1995;22:85.
307 Ramberg P, Lindhe J, Dahlen G, et al. The influence of gingival inflammation on de novo plaque formation. J Clin Periodontol. 1994;21:51.
308 Rams TE, Feik D, Slots J. Staphylococci in human periodontal diseases. Oral Microbiol Immunol. 1990;5:29.
309 Ramsey MM, Whiteley M. Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proc Natl Acad Sci U S A. 2009;106:1578.
310 Reed WP, Williams RC. Bacterial adherence: first step in pathogenesis of certain infections. J Chronic Dis. 1978;31:67.
311 Renvert S, Dahlen G, Wikstrom M. Treatment of periodontal disease based on microbiological diagnosis. Relation between microbiological and clinical parameters during 5 years. J Periodontol. 1996;67:562.
312 Renvert S, Dahlen G, Wikstrom M. The clinical and microbiological effects of non-surgical periodontal therapy in smokers and non-smokers. J Clin Periodontol. 1998;25:153.
313 Renvert S, Wikstrom M, Dahlen G, et al. Effect of root debridement on the elimination of Actinobacillus actinomycetemcomitans and Bacteroides gingivalis from periodontal pockets. J Clin Periodontol. 1990;17:345.
314 Renvert S, Wikstrom M, Dahlen G, et al. On the inability of root debridement and periodontal surgery to eliminate Actinobacillus actinomycetemcomitans from periodontal pockets. J Clin Periodontol. 1990;17:351.
315 Rickard AH, Palmer RJJr, Blehert DS, et al. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol Microbiol. 2006;60:1446.
316 Rimondini L, Fare S, Brambilla E, et al. The effect of surface roughness on early in vivo plaque colonization on titanium. J Periodontol. 1997;68:556.
317 Roberts FA, Darveau RP. Beneficial bacteria of the periodontium. Periodontol 2000. 2002;30:40.
318 Rosen G, Genzler T, Sela MN. Coaggregation of Treponema denticola with Porphyromonas gingivalis and Fusobacterium nucleatum is mediated by the major outer sheath protein of Treponema denticola. FEMS Microbiol Lett. 2008;289:59.
319 Rotimi VO, Duerden BI. The development of the bacterial flora in normal neonates. Journal of Medical Microbiology. 1981;14:51.
320 Rudney JD, Chen R, Sedgewick GJ. Intracellular Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in buccal epithelial cells collected from human subjects. Infect Immun. 2001;69:2700.
321 Rudney JD, Chen R, Sedgewick GJ. Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, and Tannerella forsythensis are components of a polymicrobial intracellular flora within human buccal cells. J Dent Res. 2005;84:59.
322 Russel AL. Epidemiology of periodontal disease. Int Dent J. 1967;17:282.
323 Rutter PR, Vincent B. Physicochemical interactions of the substratum, microorganisms and the fluid phase. In: Marshall KC, editor. Microbial adhesion and aggregation. Berlin: Springer Verlag; 1984:21.
324 Ryder MI. An update on HIV and periodontal disease. J Periodontol. 2002;73:1071.
325 Saadi AT, Blackwell CC, Essery SD, et al. Developmental and environmental factors that enhance binding of Bordatella pertussis to human epithelial cells in relation to sudden infant death syndrome (SIDS). FEMS Immunol Med Microbiol. 1996;16:51.
326 Saglie FR, Carranza FAJr, Newman MG, et al. Identification of tissue-invading bacteria in human periodontal disease. J Periodontal Res. 1982;17:452.
327 Saglie FR, Marfany A, Camargo P. Intragingival occurrence of Actinobacillus actinomycetemcomitans and Bacteroides gingivalis in active destructive periodontal lesions. J Periodontol. 1988;59:259.
328 Saito A, Inagaki S, Kimizuka R, et al. Fusobacterium nucleatum enhances invasion of human gingival epithelial and aortic endothelial cells by Porphyromonas gingivalis. FEMS Immunol Med Microbiol. 2008;54:349.
329 Samaranayake LP, Keung LW, Jin L. Oral mucosal fungal infections. Periodontol 2000. 2009;49:39.
330 Samonis G, Mantadakis E, Maraki S. Orofacial viral infections in the immunocompromised host. Oncol Rep. 2000;7:1389.
331 Sanford BA, Shelokov A, Ramsay MA. Bacterial adherence to virus-infected cells: a cell culture model of bacterial superinfection. J Infect Dis. 1978;137:176.
332 Sanz M, Quirynen M. Advances in the aetiology of periodontitis. Group A consensus report of the 5th European Workshop in Periodontology. J Clin Periodontol. 2005;32(Suppl 6):54.
333 Sbordone L, Barone A, Ciaglia RN, et al. Longitudinal study of dental implants in a periodontally compromised population. J Periodontol. 1999;70:1322.
334 Scannapieco FA. Saliva-bacterium interactions in oral microbial ecology. Crit Rev Oral Biol Med. 1994;5:203.
335 Schaeffer LM, Schmidt ML, Demuth DR. Induction of Aggregatibacter actinomycetemcomitans leukotoxin expression by IS1301 and orfA. Microbiology. 2008;154:528.
336 Schei O, Waerhaug J, Loval A. Alveolar bone loss as related to oral hygiene and age. J Periodontol. 1959;30:7.
337 Schneider LC, Schneider AE. Diagnosis of oral ulcers. Mt Sinai J Med. 1998;65:383.
338 Schreiner HC, Sinatra K, Kaplan JB, et al. Tight-adherence genes of Actinobacillus actinomycetemcomitans are required for virulence in a rat model. Proc Natl Acad Sci U S A. 2003;100:7295.
339 Schroeder HE, De Boever J. The structure of microbial dental plaque. In: McHugh WD, editor. Dental plaque. Edinburgh: Livingstone; 1970:49.
340 Schuman P, Ohmit SE, Sobel JD, et al. Oral lesions among women living with or at risk for HIV infection. HIV Epidemiology Research Study (HERS) Group. Am J Med. 1998;104:559.
341 Scully C, Felix DH. Oral medicine–update for the dental practitioner. Aphthous and other common ulcers. Br Dent J. 2005;199:259.
342 Sheiham A. Periodontal disease and oral cleanliness in tobacco smokers. J Periodontol. 1971;42:259.
343 Shibli JA, Melo L, Ferrari DS, et al. Composition of supra- and subgingival biofilm of subjects with healthy and diseased implants. Clin Oral Implants Res. 2008;19:975.
344 Simonsson T. Aspects of dental plaque formation with special reference to colloid-chemical phenomena. Swed Dent J Suppl. 1989;58:1.
345 Simonsson T, Ronstrom A, Rundegren J, et al. Rate of plaque formation–some clinical and biochemical characteristics of “heavy” and “light” plaque formers. Scand J Dent Res. 1987;95:97.
346 Siqueira WL, Oppenheim FG. Small molecular weight proteins/peptides present in the in vivo formed human acquired enamel pellicle. Arch Oral Biol. 2009;54:437.
347 Siqueira WL, Zhang W, Helmerhorst EJ, et al. Identification of protein components in in vivo human acquired enamel pellicle using LC-ESI-MS/MS. J Proteome Res. 2007;6:2152.
348 Sjodin B, Crossner CG, Unell L, et al. A retrospective radiographic study of alveolar bone loss in the primary dentition in patients with localized juvenile periodontitis. J Clin Periodontol. 1989;16:124.
349 Skopek RJ, Liljemark WF, Bloomquist CG, et al. Dental plaque development on defined streptococcal surfaces. Oral Microbiol Immunol. 1993;8:16.
350 Slaney JM, Gallagher A, duse-Opoku J, et al. Mechanisms of resistance of Porphyromonas gingivalis to killing by serum complement. Infect Immun. 2006;74:5352.
351 Sliepen I, Hofkens J, Van EM, et al. Aggregatibacter actinomycetemcomitans adhesion inhibited in a flow cell. Oral Microbiol Immunol. 2008;23:520.
352 Sliepen I, Van Damme J, Van Essche M, et al. Microbial interactions influence inflammatory host cell responses. J Dent Res. 2009.
353 Slots J. The predominant cultivable microflora of advanced periodontitis. Scand J Dent Res. 1977;85:114.
354 Slots J. Subgingival microflora and periodontal disease. J Clin Periodontol. 1979;6:351.
355 Slots J. Herpesviruses in periodontal diseases. Periodontol 2000. 2005;38:33.
356 Slots J. Oral viral infections of adults. Periodontol 2000. 2009;49:60.
357 Slots J, Gibbons RJ. Attachment of Bacteroides melaninogenicus subsp. asaccharolyticus to oral surfaces and its possible role in colonization of the mouth and of periodontal pockets. Infect Immun. 1978;19:254.
358 Slots J, Listgarten MA. Bacteroides gingivalis, Bacteroides intermedius and Actinobacillus actinomycetemcomitans in human periodontal diseases. J Clin Periodontol. 1988;15:85.
359 Slots J, Rams TE. New views on periodontal microbiota in special patient categories. J Clin Periodontol. 1991;18:411.
360 Socransky SS, Gibbons RJ, Dale AC, et al. The microbiota of the gingival crevice area of man. I. Total microscopic and viable counts of specific microorganisms. Arch Oral Biol. 1953:275.
361 Socransky SS, Haffajee AD. Microbial mechanisms in the pathogenesis of destructive periodontal diseases: a critical assessment. J Periodontal Res. 1991;26:195.
362 Socransky SS, Haffajee AD. The bacterial etiology of destructive periodontal disease: current concepts. J Periodontol. 1992;63:322.
363 Socransky SS, Haffajee AD. The nature of periodontal diseases. Ann Periodontol. 1997;2:3.
364 Socransky SS, Haffajee AD, Cugini MA, et al. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134.
365 Socransky SS, Haffajee AD, Goodson JM, et al. New concepts of destructive periodontal disease. J Clin Periodontol. 1984;11:21.
366 Socransky SS, Haffajee AD, Smith GL, et al. Difficulties encountered in the search for the etiologic agents of destructive periodontal diseases. J Clin Periodontol. 1987;14:588.
367 Socransky SS, Manganiello AD, Propas D, et al. Bacteriological studies of developing supragingival dental plaque. J Periodontal Res. 1977;12:90.
368 Socransky SS, Manganiello SD. The oral microbiota of man from birth to senility. J Periodontol. 1971;42:485.
369 Socransky SS, Smith C, Haffajee AD. Subgingival microbial profiles in refractory periodontal disease. J Clin Periodontol. 2002;29:260.
370 Stenderup A. Oral mycology. Acta Odontol Scand. 1990;48:3.
371 Stingu CS, Eschrich K, Rodloff AC, et al. Periodontitis is associated with a loss of colonization by Streptococcus sanguinis. J of Med Microbiol. 2008;57:495.
372 Stoltenberg JL, Osborn JB, Pihlstrom BL, et al. Association between cigarette smoking, bacterial pathogens, and periodontal status. J Periodontol. 1993;64:1225.
373 Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317.
374 Sumida S, Ishihara K, Kishi M, et al. Transmission of periodontal disease-associated bacteria from teeth to osseointegrated implant regions. Int J Oral Maxillofac Implants. 2002;17:696.
375 Syed SA, Loesche WJ. Bacteriology of human experimental gingivitis: effect of plaque age. Infect Immun. 1978;21:821.
376 Sztajer H, Lemme A, Vilchez R, et al. Autoinducer-2-regulated genes in Streptococcus mutans UA159 and global metabolic effect of the luxS mutation. J Bacteriol. 2008;190:401.
377 Takashiba S, Naruishi K. Gene polymorphisms in periodontal health and disease. Periodontol 2000. 2006;40:94.
378 Takenaka S, Trivedi HM, Corbin A, et al. Direct visualization of spatial and temporal patterns of antimicrobial action within model oral biofilms. Appl Environ Microbiol. 2008;74:1869.
379 Tanner AC, Haffer C, Bratthall GT, et al. A study of the bacteria associated with advancing periodontitis in man. J Clin Periodontol. 1979;6:278.
380 Tappuni AR, Challacombe SJ. Distribution and isolation frequency of eight streptococcal species in saliva from predentate and dentate children and adults. J Dent Res. 1993;72:31.
381 Taylor DC, Clancy RL, Cripps AW, et al. An alteration in the host-parasite relationship in subjects with chronic bronchitis prone to recurrent episodes of acute bronchitis. Immunol Cell Biol. 1994;72:143.
382 Teles RP, Haffajee AD, Socransky SS. Microbiological goals of periodontal therapy. Periodontol 2000. 2006;42:180.
383 Teughels W, Kinder Haake SA, Sliepen I, et al. Bacteria interfere with A. actinomycetemcomitans colonization. J Dent Res. 2007;87:611.
384 Teughels W, Newman MG, Coucke W, et al. Guiding periodontal pocket recolonization: a proof of concept. J Dent Res. 2007;86:1078.
385 Teughels W, Sliepen I, Quirynen M, et al. Human cytomegalovirus enhances A. actinomycetemcomitans adherence to cells. J Dent Res. 86, 2007.
386 Teughels W, Van EM, Sliepen I, et al. Probiotics and oral healthcare. Periodontol 2000. 2008;48:111.
387 Theilade E, Wright WH, Jensen SB, et al. Experimental gingivitis in man. II. A longitudinal clinical and bacteriological investigation. J Periodontal Res. 1966;1:1.
388 Ting M, Contreras A, Slots J. Herpesvirus in localized juvenile periodontitis. J Periodontal Res. 2000;35:17.
389 Tonetti MS, Pini-Prato G, Cortellini P. Effect of cigarette smoking on periodontal healing following GTR in infrabony defects. A preliminary retrospective study. J Clin Periodontol. 1995;22:229.
390 Tong H, Chen W, Merritt J, et al. Streptococcus oligofermentans inhibits Streptococcus mutans through conversion of lactic acid into inhibitory H2O2: a possible counteroffensive strategy for interspecies competition. Mol Microbiol. 2007;63:872.
391 Truper HG, De Clari L. Taxonomic note: necessary correction of specific epithets formed as substantives (nouns) “in apposition,”. Int J Syst Bacteriol. 1997;47:908.
392 Umeda M, Chen C, Bakker I, et al. Risk indicators for harboring periodontal pathogens. J Periodontol. 1998;69:1111.
393 Vaahtoniemi LH, Raisanen S, Stenfors LE. Attachment of bacteria to oral epithelial cells in vivo: a possible correlation to gingival health status. J Periodontal Res. 1993;28:308.
394 van der Mei HC, Rustema-Abbing M, Bruinsma GM, et al. Influence of weight on removal of co-adhering bacteria from salivary pellicles by different modes of brushing. Caries Res. 2004;38:85.
395 van der Reijden WA, Dellemijn-Kippuw N, Stijne-van Nes AM, et al. Mutans streptococci in subgingival plaque of treated and untreated patients with periodontitis. J Clin Periodontol. 2001;28:686.
396 Van der Velden U. Purpose and problems of periodontal disease classification. Periodontol 2000. 2005;39:13.
397 Van Hoogmoed CG, Geertsema-Doornbusch G, Teughels W, et al. Reduction of periodontal pathogens adhesion by antagonistic strains. Oral Microbiol Immunol. 2007.
398 van Loosdrecht MC, Norde W, Zehnder AJ. Physical chemical description of bacterial adhesion. J Biomater Appl. 1990;5:91.
399 van Steenberghe D, Jacobs R, Desnyder M, et al. The relative impact of local and endogenous patient-related factors on implant failure up to the abutment stage. Clin Oral Implants Res. 2002;13:617.
400 van Winkelhoff AJ, Bosch-Tijhof CJ, Winkel EG, et al. Smoking affects the subgingival microflora in periodontitis. J Periodontol. 2001;72:666.
401 van Winkelhoff AJ, Boutaga K. Transmission of periodontal bacteria and models of infection. J Clin Periodontol. 2005;32(Suppl 6):16.
402 van Winkelhoff AJ, Herrera D, Oteo A, et al. Antimicrobial profiles of periodontal pathogens isolated from periodontitis patients in The Netherlands and Spain. J Clin Periodontol. 2005;32:893.
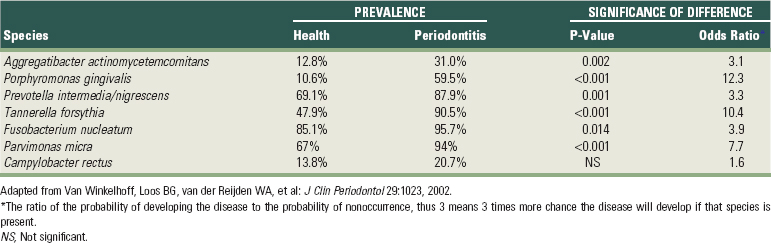
403 van Winkelhoff AJ, Loos BG, van der Reijden WA, et al. Porphyromonas gingivalis, Bacteroides forsythus and other putative periodontal pathogens in subjects with and without periodontal destruction. J Clin Periodontol. 2002;29:1023.
404 Vartoukian SR, Palmer RM, Wade WG. Diversity and morphology of members of the phylum “synergistetes” in periodontal health and disease. Appl Environ Microbiol. 2009;75:3777.
405 Vianna ME, Conrads G, Gomes BP, et al. Identification and quantification of archaea involved in primary endodontic infections. J Clin Microbiol. 2006;44:1274.
406 Vickerman MM, Brossard KA, Funk DB, et al. Phylogenetic analysis of bacterial and archaeal species in symptomatic and asymptomatic endodontic infections. J Med Microbiol. 2007;56:110.
407 Wade WG, Addy M. In vitro activity of a chlorhexidine-containing mouthwash against subgingival bacteria. J Periodontol. 1989;60:521.
408 Walden WC, Hentges DJ. Differential effects of oxygen and oxidation-reduction potential on the multiplication of three species of anaerobic intestinal bacteria. Appl Microbiol. 1975;30:781.
409 Walker C, Sedlacek MJ. An in vitro biofilm model of subgingival plaque. Oral Microbiol Immunol. 2007;22:152.
410 Walker CB. Selected antimicrobial agents: mechanisms of action, side effects and drug interactions. Periodontol 2000. 1996;10:12.
411 Wan AK, Seow WK, Purdie DM, et al. Oral colonization of Streptococcus mutans in six-month-old predentate infants. J Dent Res. 2001;80:2060.
412 Wan AK, Seow WK, Purdie DM, et al. A longitudinal study of Streptococcus mutans colonization in infants after tooth eruption. J Dent Res. 2003;82:504.
413 Wan AK, Seow WK, Walsh LJ, et al. Association of Streptococcus mutans infection and oral developmental nodules in pre-dentate infants. J Dent Res. 2001;80:1945.
414 Wang Y, Chen C. Mutation analysis of the flp operon in Actinobacillus actinomycetemcomitans. Gene. 2005;351:61.
415 Weiger R, Netuschil L, von Ohle C, et al. Microbial generation time during the early phases of supragingival dental plaque formation. Oral Microbiol Immunol. 1995;10:93.
416 Weiss EI, Eli I, Shenitzki B, et al. Identification of the rhamnose-sensitive adhesin of Capnocytophaga ochracea ATCC 33596. Arch Oral Biol. 1990;35(Suppl):127S.
417 Weiss EI, London J, Kolenbrander PE, et al. Characterization of monoclonal antibodies to fimbria-associated adhesins of Bacteroides loescheii PK1295. Infect Immun. 1988;56:219.
418 Weiss EI, London J, Kolenbrander PE, et al. Localization and enumeration of fimbria-associated adhesins of Bacteroides loescheii. J Bacteriol. 1988;170:1123.
419 Wennstrom JL, Heijl L, Dahlen G, et al. Periodic subgingival antimicrobial irrigation of periodontal pockets (I). Clinical observations. J Clin Periodontol. 1987;14:541.
420 Weyant RJ, Burt BA. An assessment of survival rates and within-patient clustering of failures for endosseous oral implants. J Dent Res. 1993;72:2.
421 Whittaker CJ, Clemans DL, Kolenbrander PE. Insertional inactivation of an intrageneric coaggregation-relevant adhesin locus from Streptococcus gordonii DL1 (Challis). Infect Immun. 1996;64:4137.
422 Wickstrom C, Svensater G. Salivary gel-forming mucin MUC5B–a nutrient for dental plaque bacteria. Oral Microbiol Immunol. 2008;23:177.
423 Williams P. Role of the cell envelope in bacterial adaptation to growth in vivo in infections. Biochimie. 1988;70:987.
424 Wilson M. An introduction to the human-microbe symbiosis. In: Wilson M, editor. Microbial inhabitants of humans. New York: Cambridge University Press; 2005:1.
425 Winkel EG, Abbas F, van d V, et al. Experimental gingivitis in relation to age in individuals not susceptible to periodontal destruction. J Clin Periodontol. 1987;14:499.
426 Wohl DA, Zeng D, Stewart P, et al. Cytomegalovirus viremia, mortality, and end-organ disease among patients with AIDS receiving potent antiretroviral therapies. J Acquir Immune Defic Syndr. 2005;38:538.
427 Wolff L, Dahlen G, Aeppli D. Bacteria as risk markers for periodontitis. J Periodontol. 1994;65:498.
428 Wood SR, Kirkham J, Marsh PD, et al. Architecture of intact natural human plaque biofilms studied by confocal laser scanning microscopy. J Dent Res. 2000;79:21.
429 Ximenez-Fyvie LA, maguer-Flores A, Jacobo-Soto V, et al. Subgingival microbiota of periodontally untreated Mexican subjects with generalized aggressive periodontitis. J Clin Periodontol. 2006;33:869.
430 Ximenez-Fyvie LA, maguer-Flores A, Jacobo-Soto V, et al. Description of the subgingival microbiota of periodontally untreated Mexican subjects: chronic periodontitis and periodontal health. J Periodontol. 2006;77:460.
431 Xu KD, McFeters GA, Stewart PS. Biofilm resistance to antimicrobial agents. Microbiology. 2000;146(Pt 3):547.
432 Yao Y, Berg EA, Costello CE, et al. Identification of protein components in human acquired enamel pellicle and whole saliva using novel proteomics approaches. J Biol Chem. 2003;278:5300.
433 Yokoyama M, Hinode D, Yoshioka M, et al. Relationship between Campylobacter rectus and periodontal status during pregnancy. Oral Microbiol Immunol. 2008;23:55.
434 Zambon JJ. Periodontal diseases: microbial factors. Ann Periodontol. 1996;1:879.
435 Zambon JJ, Grossi SG, Machtei EE, et al. Cigarette smoking increases the risk for subgingival infection with periodontal pathogens. J Periodontol. 1996;67:1050.
436 Zee KY, Samaranayake LP, Attstrom R. Predominant cultivable supragingival plaque in Chinese “rapid” and “slow” plaque formers. J Clin Periodontol. 1996;23:1025.
437 Zee KY, Samaranayake LP, Attstrom R. Scanning electron microscopy of microbial colonization of ‘rapid’ and ‘slow’ dental-plaque formers in vivo. Arch Oral Biol. 1997;42:735.
438 Zhang G, Chen R, Rudney JD. Streptococcus cristatus attenuates Fusobacterium nucleatum-induced interleukin-8 expression in oral epithelial cells. J Periodontal Res. 2008;43:408.