Ribonucleic Acid
Introduction
Transcription is defined as the synthesis of a ribonucleic acid (RNA) molecule using deoxyribonucleic acid (DNA) as a template
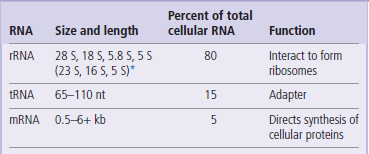
Transcription is a series of complicated enzymatic processes that result in the transfer of the genetic information stored in double-stranded DNA into a single-stranded RNA molecule that will be used by the cell to direct the synthesis of its proteins, a process known as translation. There are three general classes of RNA molecules found in prokaryotic and eukaryotic cells: ribosomal RNA (rRNA), transfer RNA (tRNA), and messenger RNA (mRNA). Each class has a distinctive size and function (Table 33.1), described by its sedimentation rate in an ultracentrifuge (S, Svedberg units) or its number of bases (nt, nucleotides, or kb, kilobases). Prokaryotes have the same three general classes of RNA as eukaryotes, but sizes and structural features differ:
 ribosomal RNA (rRNA) from prokaryotes consists of three different sizes of RNA, while rRNA from eukaryotes consists of four different sizes of RNA. These RNAs interact with each other, and with proteins, to form a ribosome that provides the basic machinery on which protein synthesis takes place.
ribosomal RNA (rRNA) from prokaryotes consists of three different sizes of RNA, while rRNA from eukaryotes consists of four different sizes of RNA. These RNAs interact with each other, and with proteins, to form a ribosome that provides the basic machinery on which protein synthesis takes place.
 transfer RNAs (tRNAs) consist of a class of RNAs that are 65–110 nt in length; they function as amino acid carriers and as recognition molecules that identify the mRNA nucleotide sequence and translate that sequence into the amino acid sequence of proteins.
transfer RNAs (tRNAs) consist of a class of RNAs that are 65–110 nt in length; they function as amino acid carriers and as recognition molecules that identify the mRNA nucleotide sequence and translate that sequence into the amino acid sequence of proteins.
 messenger RNAs (mRNAs) represent the most heterogeneous class of RNAs found in cells. mRNAs generally range in size from 500 nt to ~6 kb (some rare but important mRNAs are >100 kb). mRNAs are carriers of genetic information, defining the sequence of all proteins in the cell; they are the ‘working copy’ of the genome.
messenger RNAs (mRNAs) represent the most heterogeneous class of RNAs found in cells. mRNAs generally range in size from 500 nt to ~6 kb (some rare but important mRNAs are >100 kb). mRNAs are carriers of genetic information, defining the sequence of all proteins in the cell; they are the ‘working copy’ of the genome.
In order to understand the complex series of events that lead to the production of these three classes of RNA, this chapter is divided into five parts. The first part deals with the molecular anatomy of the major types of RNAs found in prokaryotic and eukaryotic cells; by understanding the chemical nature of the final products of transcription, you will be better prepared to understand the steps involved in generating these molecules. The second part describes the main enzymes involved in transcription, and their specificities. The third part describes the three steps (initiation, elongation, and termination) required to produce a protein. In the fourth section, the modifications that are made to the primary products of transcription (post-transcriptional processing) are described. This information is expanded in Chapter 34. The final section describes briefly how cells regulate gene expression at the RNA level.
Molecular anatomy of ribonucleic acid molecules
In contrast to DNA, RNAs are mostly single stranded, and contain uracil instead of thymine
In general, the RNAs produced by prokaryotic and eukaryotic cells are single-stranded nucleic acid molecules that consist of adenine, guanine, cytosine, and uracil nucleotides joined to one another by phosphodiester linkages. The start of an RNA molecule is known as its 5′ end, and the termination of the RNA is its 3′ end. Even though most RNAs are single-stranded, they fold back on themselves. Therefore, they exhibit extensive secondary structure, including intramolecular double-stranded regions that are important to their function. These secondary structures, one of the most common of which is called a hairpin loop (Fig. 33.1), are the product of intramolecular base pairing that occurs between complementary nucleotides within a single RNA molecule.
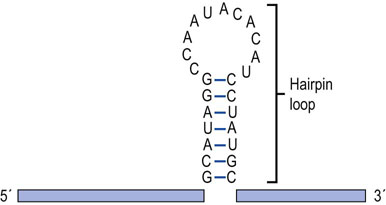
Fig. 33.1 RNA hairpin loop.
RNA can form secondary structures called hairpin loops. These structures form when complementary bases within an individual RNA share hydrogen bonds and form base pairs. Hairpin loops are known to be important in the regulation of transcription in both eukaryotic and prokaryotic cells.
rRNAs: the ribosomal RNAs
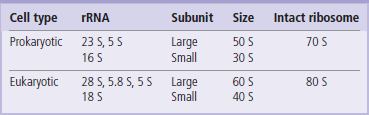
The eukaryotic rRNAs are initially synthesized as a single RNA transcript with a size of 45 S and about 13 kb long. This large primary transcript is processed into 28 S, 18 S, 5.8 S, and 5 S rRNAs (~3 kb, 1.5 kb, 160 and 120 nt, respectively). The 28 S, 5.8 S, and 5 S rRNAs associate with ribosomal proteins to form the large ribosomal subunit. The 18 S rRNA associates with different proteins to form the small ribosomal subunit. The large ribosomal subunit with its proteins and RNA has a characteristic size of 60 S; the small ribosomal subunit has a size of 40 S. These two subunits interact to form a functional 80 S ribosome (see Chapter 34). Prokaryotic rRNAs interact in a similar fashion to form these ribosomal subunits but have a slightly smaller size, reflecting the difference in prokaryotic and eukaryotic rRNA transcript size (Table 33.2).
tRNA: the molecular cloverleaf
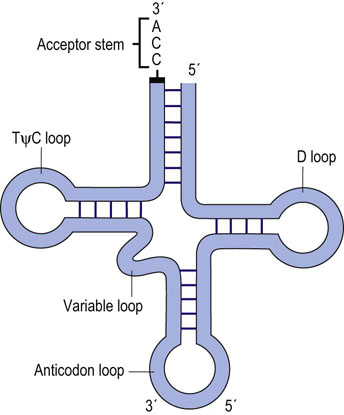
Prokaryotic and eukaryotic tRNAs are similar in both size and structure. They exhibit extensive secondary structure and contain several modified ribonucleotides that are derived from the normal four ribonucleotides. All tRNAs have a similar fold, with four distinct loops that have been described as a cloverleaf (Fig. 33.2). The D loop contains several modified bases, including methylated cytosine and dihydrouridine (D), for which the loop is named. The anticodon loop is the structure responsible for recognition of the complementary codon of an mRNA molecule: specific interaction of an anticodon of the tRNA with the appropriate codon in the mRNA is due to base pairing between these two complementary trinucleotide sequences. A variable loop, 3–21 nt in length, exists in most tRNAs but its function is unknown. Finally, there is a TψC loop, which contains a modified base, pseudouridine (ψ). Another prominent structure found in all tRNA molecules is the acceptor stem. This structure is formed by base pairing between the nucleotides at each end of the tRNA. The last three bases found at the extreme 3′ end remain unpaired, and always have the same sequence: 5′-CCA-3′. This 3′ end of the acceptor stem is the point at which an amino acid is attached via an ester bond between the 3′-hydroxyl group of the adenosine and the carboxyl group of an amino acid in preparation for protein synthesis (see Chapter 34).
mRNA: prokaryotes and eukaryotes have dissimilar mRNAs
Prokaryotes and eukaryotes are very different kinds of organisms with dramatically different life cycles. Therefore, it is not surprising that there are differences in the structures of their genes, in their mechanisms of transcription, and in the structures of their mRNAs. In fact, we can exploit these differences with novel antibiotics that target unique portions of the prokaryotic life cycle. There are a number of major differences between prokaryotic and eukaryotic mRNAs. These will be discussed in sections below. Briefly, these differences are:
 Transcriptional units differ in structure: prokaryotic mRNAs are polycistronic, eukaryotic mRNAs are monocistronic (Fig. 33.3).
Transcriptional units differ in structure: prokaryotic mRNAs are polycistronic, eukaryotic mRNAs are monocistronic (Fig. 33.3).
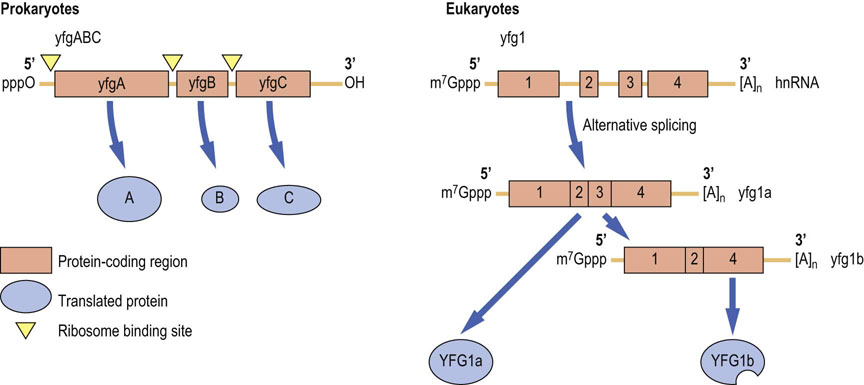
Fig. 33.3 Prototypical structures of prokaryotic (polycistronic) and eukaryotic mRNAs.
Prokaryotic mRNAs have naked ends (triphosphate at the 5′ end and hydroxyl at the 3′ end). The boxes indicate those portions of the mRNA that encode a protein. Inverted triangles indicate location of ribosome-binding sites. The three genes in this cistron are translated into three different proteins. Other prokaryotic cistrons may encode up to 10 different proteins in a single mRNA. Nascent eukaryotic mRNA transcripts (heterogeneous nuclear RNA (hnRNA)) contain both exons (boxes) and introns (lines). Eukaryotic mRNAs are protected by a 7-methylguanine nucleotide cap (m7Gppp) at the 5′ end and a polyA tail ([A]n) at the 3′ end of the mRNA. After splicing, the mature mRNAs consist only of exons plus the 5′ and 3′ UTRs. Alternatively spliced mRNAs are translated into different protein isoforms. yfg: your favorite gene; UTR: untranslated region.
 Compartmentalization of transcription and translation: prokaryotes synthesize RNA and protein in one compartment, the cellular cytoplasm, eukaryotes separate these events in the nucleus and cytoplasm.
Compartmentalization of transcription and translation: prokaryotes synthesize RNA and protein in one compartment, the cellular cytoplasm, eukaryotes separate these events in the nucleus and cytoplasm.
 Protection at their 5′ and 3′ ends: ends of prokaryotic mRNAs are naked, eukaryotic mRNAs have a 5′-cap and 3′-poly(A) tail.
Protection at their 5′ and 3′ ends: ends of prokaryotic mRNAs are naked, eukaryotic mRNAs have a 5′-cap and 3′-poly(A) tail.
 Processing of mRNAs: prokaryotic mRNAs are not processed; eukaryotic mRNAs contain introns that are spliced out.
Processing of mRNAs: prokaryotic mRNAs are not processed; eukaryotic mRNAs contain introns that are spliced out.
A major difference between prokaryotic and eukaryotic mRNAs relates to their transcriptional unit structure. In prokaryotes, transcriptional units generally contain multiple protein-coding regions (see Fig. 33.3), while in eukaryotes, each transcriptional unit generally codes for only a single protein. The polycistronic mRNAs of prokaryotes have individual start and stop codons at the beginning and end of each open reading frame, the sequence of mRNA that specifies the sequence of the polypeptide chain. Each stop codon is closely followed by another ribosome binding site and a translation start site that functions for the next open reading frame.
A second major difference between prokaryotic and eukaryotic mRNAs is the compartmentalization of the transcription and translation processes. Because prokaryotes lack a nucleus, transcription and translation are intimately coupled in the cytoplasm; prokaryotic translation is usually initiated before transcription is finished. By coupling these processes, prokaryotes increase the rate at which proteins are expressed, consistent with the relatively short life cycles of prokaryotes. In contrast, eukaryotic cells separate transcription in the nucleus from translation in the cytoplasm. Although this arrangement slows response time for protein production, it allows for much more subtle control of protein expression.
The post-transcriptional processing of mRNAs is also significantly different in prokaryotes and eukaryotes. Because of their importance, these differences will be detailed in a separate section below dealing with post-transcriptional processing of RNAs. Briefly, eukaryotes protect the 5′ and 3′ ends of mRNAs by addition of specific molecular structures (5′ cap and polyA tail) that function to reduce mRNA turnover. Also, eukaryotic genes contain introns, untranslated sequences that are present in nascent transcripts and must be spliced out to produce mature mRNAs.
Ribonucleic acid polymerases
RNA polymerases are large multimeric enzymes that transcribe defined segments of DNA into RNA with a high degree of selectivity and specificity
The enzymes responsible for the synthesis of RNA are called RNA polymerases (RNAPol). In contrast to DNA polymerases (Chapter 32), RNA polymerases do not require a primer to initiate RNA synthesis. The RNA polymerases generally consist of two high-molecular-weight subunits and several smaller subunits, all of which are necessary for accurate transcription. Prokaryotes contain a single RNA polymerase that synthesizes all RNAs; however, eukaryotes contain three different RNA polymerases, termed RNA polymerase I, II, and III.
Each polymerases specialize in transcription of one class of RNA
 RNAPol I transcribes ribosomal RNAs. The rRNAs are produced from a single transcriptional unit that is subsequently processed to produce the 18 S, 28 S, 5.8 S and 5 S rRNAs.
RNAPol I transcribes ribosomal RNAs. The rRNAs are produced from a single transcriptional unit that is subsequently processed to produce the 18 S, 28 S, 5.8 S and 5 S rRNAs.
 RNAPol II transcribes most genes within a eukaryotic cell, including all protein-coding genes that yield mRNA. RNAPol II is exquisitely sensitive to α-amanitin, a potent and toxic transcription inhibitor found in some poisonous mushrooms.
RNAPol II transcribes most genes within a eukaryotic cell, including all protein-coding genes that yield mRNA. RNAPol II is exquisitely sensitive to α-amanitin, a potent and toxic transcription inhibitor found in some poisonous mushrooms.
 RNAPol III transcribes most of the small cellular RNAs, including the tRNAs.
RNAPol III transcribes most of the small cellular RNAs, including the tRNAs.
Yeast RNAPol II consists of a 12-subunit core. It exists in two forms. The first is an open form that is shaped like a cupped hand with a cleft that binds the DNA molecule and associated transcription factors near the start point of transcription. After melting or dissociation of the bound DNA strands, the complex undergoes a large structural change that closes the cleft, forming a clamp around the antisense or template strand of the DNA. Then, a specific protein (rbp4/7) binds to the base of the clamp, locking the clamp in the closed state. The closed form is no longer competent for transcript initiation but is capable of transcript elongation. The yeast RNAPol II structure appears to be an excellent model for the human enzyme. In addition, it is also a good model for the function of RNAPol I and III because the core subunits are either shared or are homologous between the various enzymes.
The bacterial RNA polymerase is similar to the eukaryotic enzyme complex, except the bacterial enzyme contains only four subunits (α2ββ′) and, unlike the eukaryotic enzyme, requires only a single general transcription factor (σ-factor) to recognize the promoter and recruit the RNA polymerase to initiate transcription.
Messenger ribonucleic acid: transcription
Transcription is a dynamic process that involves the specific interaction of enzymes with DNA to produce RNA molecules
It is convenient to divide transcription into three separate stages: initiation, elongation and termination. Transcription proceeds along the antisense or template strand, producing a complementary RNA that is identical to the sense strand of the DNA (Fig. 33.4), except that the thymine residues in DNA are substituted by uracil residues in RNA.
Initiation
Initiation involves the site-specific interaction of the RNA polymerase with DNA
Because most genomic DNA does not encode proteins, identification of transcription start sites is crucial to obtain desired mRNAs. Special sequences termed promoters recruit the RNA polymerase to the transcription start site (Fig. 33.5). Promoters are usually located in front (upstream) of the gene that is to be transcribed. However, RNAPol polymerase III promoters are actually located within the gene.
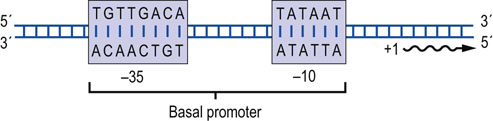
Fig. 33.5 Prokaryotic promoters.
Promoters in prokaryotic genes are located immediately upstream of the transcription start site. Two common conserved regions have been identified that are found at −35 and −10, respectively. The consensus sequences in these regions are shown. Position +1 indicates the first nucleotide that will be transcribed into RNA. The TATA box at –10 is a common AT-rich promoter element.
Prokaryotic genes generally contain simple promoters. Promoters are generally rich in adenine (A) and thymine (T). The presence of these nucleotides facilitates separation of the two DNA strands, because hydrogen bonding between A-T base pairs is weaker than between G-C base pairs. Comparisons of large numbers of prokaryotic promoters have identified two common conserved regions. These are located about 10 nucleotides and 35 nucleotides upstream (–10 and –35 sequences, respectively) from the transcription start site (see Fig. 33.5). The –10 sequence is known as the TATA box. This sequence binds the prokaryotic general transcription factor (σ-factor) that interacts and recruits the RNA polymerase to the promoter. Strong promoters tend to match the consensus sequence shown in Figure 33.5; sequences of weaker promoters differ from the consensus sequence and bind the σ-factor and the RNA polymerase less tightly.
In eukaryotic RNAPol II promoters, regulatory elements (specific short DNA sequences) termed upstream activation sequences (UASes), enhancers, repressors, CAATT and TATA box sequences are spread over several hundred to several thousand nucleotides. Individual transcription factors (activators or repressors) recognize and bind to these UASes. The control of initiation and the regulation of gene expression are outlined in detail in Chapter 34.
Elongation
Elongation is the process by which single nucleotides are added to the growing RNA chain
In prokaryotes, elongation is a relatively simple process. Ribonucleotides bind to an entry site on the RNA polymerase. If the incoming ribonucleotide matches the next base on the DNA template, the incoming ribonucleotide is transferred into the polymerase active site and a new phosphodiester bond is formed. If it does not match, the ribonucleotide is released and the process repeated until the correct ribonucleotide is found. After the formation of the phosphodiester bond, the RNA polymerase translocates along the template DNA strand. It is thought that the RNA polymerase accomplishes this by oscillating a small helical region of the RNA polymerase molecule between straight and bent conformations, permitting the polymerase to ratchet about 3 Å (=1 nucleotide step) along the antisense strand. After translocation, a new nucleotide is added.
In eukaryotes, after RNA polymerase II initiates transcription, a pair of negative elongation regulatory factors (NELF and DSIF) trap the RNA polymerase in the starting position. An RNA–protein complex, termed P-TEFb, is a kinase that phosphorylates these two inhibitory molecules, thereby releasing the RNA polymerase to continue RNA synthesis.
Vesicular stomatitis virus (VSV) and HIV produce viral proteins that stabilize the RNA polymerase complex, either directly or by recruiting host factors. The HIV protein TAT (a trans-activating regulatory protein) is one of the better understood of these stabilization proteins. Upon interaction with RNA polymerase the TAT protein rapidly recruits the P-TEFb complex, resulting in increased transcription of the full-length viral RNA, at the expense of cellular RNAs.
Elongation can be a rapid process, occurring at the rate of ~40 nt per second. For elongation to occur, the double-stranded DNA must be continually unwound, so that the template strand is accessible to the RNA polymerase. DNA topoisomerases I and II, enzymes associated with the transcription complex, move along the template with the RNA polymerase, separating DNA strands so that they are accessible for RNA synthesis.
Termination
Termination of transcription is catalyzed by multiple mechanisms in both prokaryotes and eukaryotes
At the end of a transcriptional unit, the RNA polymerases terminate RNA synthesis at defined sites. Transcriptional termination mechanisms are much better understood in prokaryotes than in eukaryotes. In prokaryotes, termination occurs via one of two well-characterized mechanisms that both require the formation of hairpin loops in the RNA secondary structure (Fig. 33.6). In rho-independent, intrinsic termination, a hairpin loop is formed just upstream of a sequence of 6–8 uridine (U) residues located near the 3′ end of the transcript. The formation of this secondary structure dislodges the RNA polymerase from the DNA template, resulting in termination of RNA synthesis. In rho-dependent termination, the RNA transcript encodes a binding site for rho protein, an ATP-dependent helicase; rho binds to and travels along the RNA transcript, unwinding it from DNA. It ‘chases’ the RNA polymerase but is slower than RNAPol II. A rho termination site near the end of the transcriptional unit causes the RNAPol II to pause, allowing the rho protein to catch up and unwind the RNA:DNA duplex, displacing the RNAPol II from the template, thereby stopping transcription.
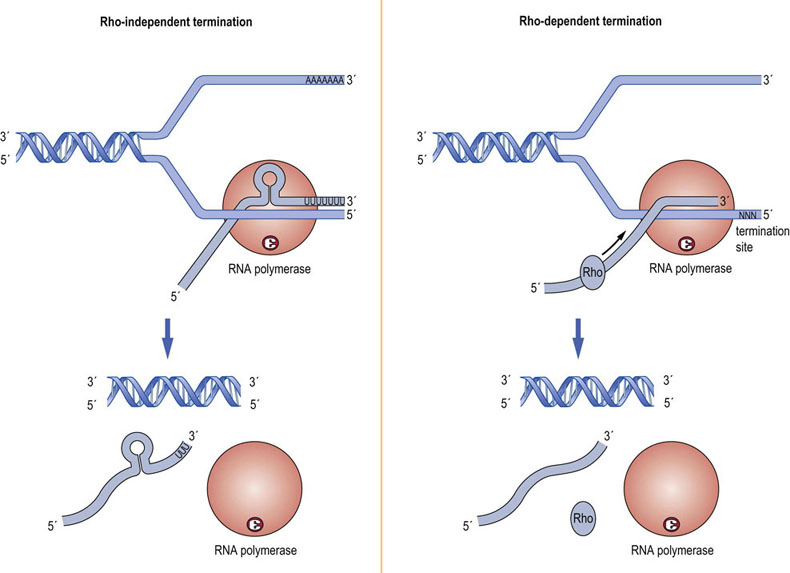
Fig. 33.6 Transcription termination in prokaryotes.
Two mechanisms of transcription termination in bacterial cells are known. Rho-independent termination relies on the formation of a secondary structure in the newly transcribed RNA to dislodge the RNA polymerase from the DNA template and stop transcription. Rho-dependent termination requires the action of the rho protein, an ATP-dependent helicase. This protein moves along the newly transcribed RNA, unwinding the RNA:DNA duplex, catching up with the RNA polymerase when it pauses at the termination site, and causing the polymerase to dissociate from the DNA template.
In eukaryotes, the three RNA polymerases employ different mechanisms to terminate transcription. RNAPol I uses a specific protein, transcript termination factor 1 (TTF1), that binds to an 18 nt terminator site located about 1000 nt downstream of the rRNA coding sequence. When RNAPol I encounters the TTF1 bound to DNA, a releasing factor catalyzes the release of the polymerase from the rRNA gene. RNAPol III uses a mechanism that is similar to bacterial ‘rho-independent’ termination; however, the length of the uridine (U) stretch is shorter and there is no requirement for a RNA secondary structure to dislodge the RNA polymerase. The mechanism of termination by RNAPol II, which transcribes most eukaryotic genes, is not well understood, in part because the RNAPol II products are immediately processed by removal of the nascent 3′ end and addition of a polyadenosine (polyA) tail.
Post-transcriptional processing of ribonucleic acids
The prokaryotic life strategy is to replicate as rapidly as possible when conditions support growth. Eukaryotes have a more controlled life strategy that invests more fully in increased regulation to achieve stable growth but limits rapid reproduction. Both strategies work well for each type of organism as evidenced by the rich diversity of life on earth; mechanisms of RNA synthesis and processing have evolved to optimize each life strategy. This is especially true for mRNAs.
Pre-rRNA and pre-tRNA
tRNAs and rRNAs are synthesized as larger precursors (pre-RNAs) that must be processed to yield mature transcripts (Fig. 33.7)
In prokaryotes a single 30 S rRNA transcript (~6.5 kb) contains specific leader and trailer regions located at the 5′ and 3′ ends of the transcript as well as one copy each of the 23 S, 16 S and 5 S rRNAs. The rRNA genes also contain a number of tRNAs that are embedded in the pre-rRNA transcript. The rRNA transcript must be processed to liberate the functional RNAs. Including each of the ribosomal RNAs in a single transcript is clearly advantageous to maintain the ratio of large-to-small ribosomal subunits.
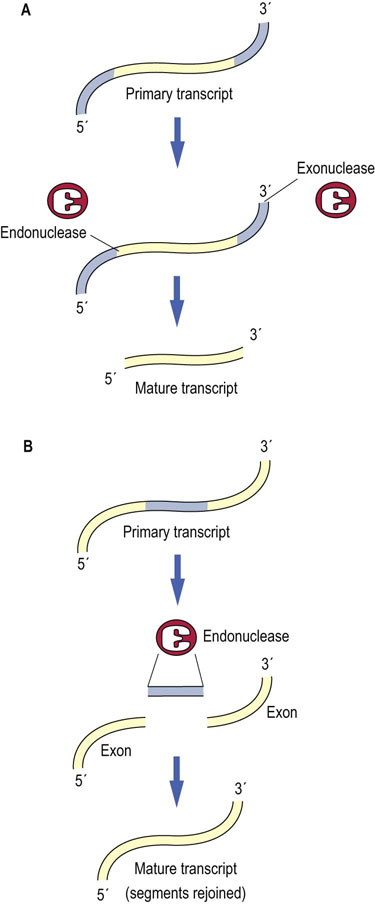
Fig. 33.7 RNA processing.
There are two general types of RNA processing events. Processing of an RNA transcript can involve (A) the removal of excess sequences by the action of endonucleases and exonucleases as in the processing of rRNA and tRNA genes, or (B) the removal of excess sequences and the rejoining of segments of the newly transcribed RNA as in splicing of mRNAs (see also Fig. 33.8).
In prokaryotes, processing of the pre-rRNA requires several RNases. Ribonuclease III (RNase III) cleaves the pre-rRNA in double-stranded regions. Such regions occur at each end of the 16 S and 23 S rRNAs and their cleavage liberates these rRNAs from the pre-rRNA transcript. The 16 S and 23 S rRNAs are further processed at the 5′ and 3′ ends; however, this trimming requires the presence of specific ribosomal proteins and occurs during ribosome assembly.
In all eukaryotes from yeast to mammals, pre-rRNA transcripts are processed in a manner similar to the prokaryotic pre-rRNA processing. Every 45 S rRNA transcript (~13.7 kb) includes a single copy of the 18 S, 5.8 S and 28 S rRNAs (in eukaryotes, the 5S rRNA is separately encoded). However, processing of the human rRNA is more complex. The pre-rRNA transcript must be cleaved at 11 different sites to generate the mature 18 S, 28 S, and 5.8 S rRNAs. Processing occurs on a huge ribonucleoprotein complex termed the processome. In addition to modifications by cleavage, the mature human rRNA contains 115 specific methyl group modifications (in most of these, the methyl group is added to the backbone ribosyl 2′-hydroxyl group, 2′-O-methyl) and 95 specific uridine to pseudouridine (ψ) conversions. These modifications are introduced into the pre-rRNA through the interaction with individual small nucleolar RNA protein complexes (snoRNPs, pronounced snorps). Each of these snoRNPs contains a unique guide RNA (~60–300 nt in length) and from one to four protein molecules. Each snoRNP is specific for a single, or at most a few, individual modification sites. The snoRNA (small ribonucleolar RNA) component of snRNPs contains structural motifs that belong to two groups, either the C/D box or the H/ACA box. These sequences direct binding of snoRNPs to sites in the pre-rRNA molecules by virtue of a complementary nucleotide stretch (~10–20 nucleotides). This correctly positions a methyl transferase (box C/D) or a pseudouridine synthase (box H/ACA) along the pre-rRNA for modification. The processome contains ≥100 snoRNAs as well as more than 100 individual proteins.
In addition to the rRNA genes, tRNAs are also synthesized in precursor form. As many as seven individual tRNAs can be synthesized from a single pre-tRNA gene. Processing of the tRNAs from the pre-tRNAs requires RNase P, which cleaves each tRNA from the pre-tRNA by a single cleavage at its 5′ end. RNAse P is an RNA–protein complex containing a 377 nucleotide RNA and a 20 kDa protein, although the protein portion is not required for enzymatic activity, i.e. the RNA is catalytic by itself. The discovery of self-splicing RNA – that is, RNA with an enzymatic activity, a ribozyme – has led to new ideas about early cellular evolution, which was originally believed to start with amino acids and proteins. It is now believed that ribonucleotides and RNA may have been the most primitive catalytic biopolymers to form on earth, providing for genetic diversity, and that DNA and proteins may have developed later. A third enzyme, RNase D, trims away the extra 3′ nucleotides from the pre-tRNAs, leaving the invariant CCA that is found at the 3′ end of every tRNA. A few tRNAs also contain introns within the anticodon loop which must be removed during processing.
Pre-mRNA processing
Prokaryotes rapidly synthesize their mRNAs and typically do not process or modify them; both the 5′ and 3′ ends of prokaryotic mRNAs are naked and unprotected. Consequently, even newly synthesized mRNAs are rapidly degraded by normal cellular RNases. This is not a problem for these rapidly growing organisms; they are able to rapidly alter mRNA synthesis for immediate protein production. After their immediate needs have been met, they subsequently degrade their mRNAs and reuse the ribonucleotides for synthesis of other mRNAs. A typical half-life of prokaryotic mRNA is about 3 minutes. In contrast, eukaryotes take special precautions to stably maintain their mRNAs for continued use. mRNA half-lives in eukaryotes range from a few minutes, for some highly regulated transcription factors, to as long as 30 hours for some long-lived transcripts.
Eukaryotic mRNAs have longer half-lives than prokaryotic mRNAs because of protective modifications at their 3′ and 5′ ends
Eukaryotes have evolved methods to protect each end of the mRNA. At the 5′ end, a unique structure termed a ‘5′ cap’ is added. The cap consists of a 7-methylguanidine residue that is attached in reverse orientation to the first nucleotide of the mRNA, i.e. by a 5′-to-5′ triphosphate linkage (m7Gppp). Most cellular exo-RNases do not have the ability to hydrolyze this cap from the mRNA, so the 5′ end is immune in their presence. At the 3′ end of nearly all eukaryotic mRNAs (with the exception of histone mRNAs), a polyadenosine track is added, termed the polyA tail. The adenosine residues are not encoded by the DNA but instead are added by the action of poly(A) polymerase using ATP as a substrate. This polyA tail is frequently >250 nucleotides in length. Although it is still susceptible to the action of exo-RNases, the presence of the polyA tail significantly reduces the turnover of mRNA, thereby increasing its lifetime. The presence of the polyA tail has historically been used to isolate mRNA from eukaryotic cells.
The spliceosome joins exons from pre-mRNA to form a mature mRNA
In the more complicated post-transcriptional processing of eukaryotic mRNAs, sequences called introns (intravening sequences) are removed from the primary transcript (pre-RNA, a form of hnRNA) and the remaining segments, termed exons (expressed sequences), are ligated to form a functional RNA. This process involves a large complex of proteins and auxiliary RNAs called small nuclear RNAs (snRNAs), which interact to form a spliceosome. The function of the five snRNAs (U1, U2, U4, U5, U6) in the spliceosome is to help position reacting groups within the substrate mRNA molecule, so that the introns can be removed and the appropriate exons can be spliced together precisely (Table 33.3). The snRNAs accomplish this task by binding, through base-pairing interactions, with the sites on the mRNA that represent intron/exon boundaries.
Table 33.3
The function of small nuclear RNAs (snRNAs) in the splicing of mRNAs
| snRNA | Size | Function |
| U1 | 165 nt | Binds the 5′ exon/intron boundary |
| U2 | 185 nt | Binds the branch site on the intron |
| U4 | 145 nt | Helps assemble the spliceosome |
| U5 | 116 nt | Binds the 3′ intron/exon boundary |
| U6 | 106 nt | Displaces U1 after first rearrangement |
The removal of an intron and rejoining of two exons can be considered to occur in three steps (Fig. 33.8). The first step involves the binding of the U1 snRNP to the exon/intron boundary at the 5′ end of the intron, along with the binding of the U2 snRNP to a target adenosine nucleotide, usually found about 30 nt from the 3′ end of the intron, Following binding of the U4/U5/U6 snRNP complex just upstream of the 5′ splice site, the intron loops back on itself positioning the ends of the introns in the correct orientation. In this process, the U6 snRNP displaces the U1 snRNP from the mRNA. Subsequently, a transesterification reaction between the 2′-hydroxyl of the target adenosine with the phosphodiester bond of the intron's 5′-guanosine residue breaks the upstream splice site and forms a branched chain structure in which the target adenine has 2′-, 3′-, and 5′-phosphate groups. The looped structure of the intron is similar in appearance to a cowboy's lariat. Following a subsequent physical rearrangement which releases the U4 snRNP, a second transesterification reaction ligates the 3′ end of exon 1 with the 5′ end of exon 2. The splicosome then disassembles releasing the lariat structure, which is degraded and the mature mRNA is ready for further processing, including addition of the M7G cap and polyA tail, and nuclear export.
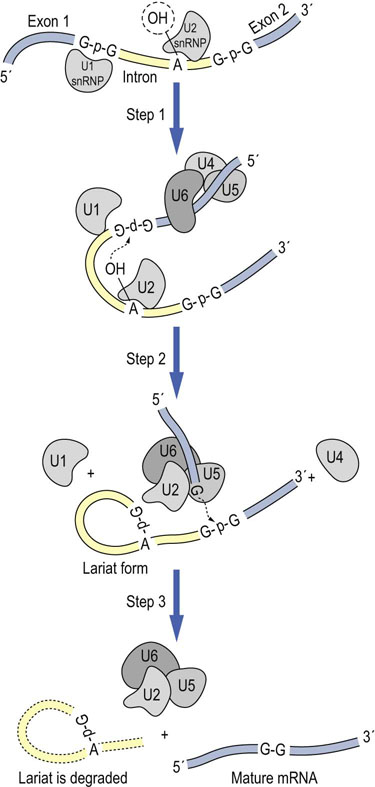
Fig. 33.8 RNA splicing.
RNA splicing is a multistep process catalyzed by ribonucleoprotein complexes, “simplified” in the above diagram. In a transesterification reaction, the phosphate bond of a guanosine residue at the 5′ exon/intron boundary is broken and joined to the 2′-OH of an adenine residue located in the middle of the intron. In a later step, the phosphate bond at the 3′ intron/exon boundary is first cleaved and then the two exons are spliced together by reformation of a phosphodiester bond between the nucleotides at either end of the exons. The intron is eliminated in the form of a lariat structure, cyclized through a 2′,3′5′-phosporylated adenosine residue. N = any nucleotide.
Alternative splicing produces multiple mature mRNAs from a single pre-mRNA transcript
Most eukaryotic mRNAs consist of multiple introns and multiple exons. If splicing were consistent, only a single mature mRNA would result from the pre-mRNAs. However, many eukaryotic genes undergo a process called alternative splicing, in which different mRNA regions are removed from the pre-mRNA, resulting in multiple mature mRNAs with different sequences (see Fig. 33.3). When these different mRNAs are translated, multiple protein isoforms are made. In humans, almost 60% of pre-mRNAs give rise to multiple mature mRNAs following alternative splicing. About 80% of these alternatively spliced mRNAs result in alternations in the encoded proteins (splicing events may also occur in the 5′ and 3′ untranslated regions). Alternative splicing can result in insertion or deletion of amino acids in the protein sequence, shifts in reading frames, or even introduction of novel stop codons. Such alternative splicing can also add or remove mRNA sequences that can alter regulatory elements affecting translation, mRNA stability or subcellular localization. In vertebrates, the introns of many ribosomal protein genes host the sequences of the small nucleolar RNAs (snoRNAs) that function in modification of specific residues in rRNAs and tRNAs; however, many introns do not host these sequences and their function remains obscure.
Editosomes modify the nucleotide sequence of mature mRNAs
Finally, in some instances, mRNAs are post-transcriptionally edited via one of several different mechanisms. These include specific C-to-U modifications catalyzed by cytosine deaminases or A-to-I modifications catalyzed by adenosine deaminases or deletion of single or multiple U residues to change the nucleotide sequence of the mRNA. These changes lead to codon differences that result in altered protein sequences that differ from the sequences encoded at the gene level (see Chapter 35). A large multiprotein complex termed the editosome is involved in this process. The snoRNAs that function in rRNA and tRNA modification also function in these RNA editing processes. These snoRNAs bind to the editing sites within the mRNAs and position the editosome with either cytosine deaminase or adenosine deaminase activity at the correct position to complete the modification.
Selective degradation or inactivation of ribonucleic acid
Interferon activates pathways that inhibit proliferation of RNA viruses
RNA viruses pose a major challenge for eukaryotic cells; however, natural defense mechanisms have evolved to limit viral infection. Because RNA viruses generally form a double-strand (ds) replicative intermediate during their life cycle, this dsRNA is a unique structure that is not generally found in eukaryotic cells. It can be recognized by dsRNA-binding proteins that will subsequently trigger responses to limit viral infection. One of these mechanisms involves the dsRNA-activated protein kinase R (PKR). When activated by binding dsRNA, this enzyme can phosphorylate and inactivate the protein translation factor, eIF2α, thereby downregulating translation of viral RNA. Likewise, when activated by dsRNA, the enzyme 2′-5′-oligoadenylate synthase polymerizes ATP into a series of short nucleotides (2′-5′-oligoadenylate, 2–5 A) that differs from the 5′-3′ structure found in normal RNA. The most active form is a trimer, pppA-2′-p-5′-A-2′-p-5′-A. The accumulation of 2–5 A inhibits viral (and host) protein translation by activating an endoribonuclease (RNase L) that indiscriminately degrades both mRNAs and rRNAs within the cell. Both genes, PKR and 2–5 A synthase, are induced by interferon, which is itself upregulated by viral infection. This results in an efficient amplification mechanism, leading to programmed cell death (apoptosis) to limit the growth and spread of the virus.
RNAis exert wide-ranging effects on gene expression
Another important process in innate cellular immunity involves specific targeting of RNA sequences for rapid degradation. This process is termed RNA interference (RNAi) (see Chapter 35) or post-transcriptional gene silencing (PTGS). While this process is thought to have evolved as a defense against double-strand forms of RNA viruses, it also functions as an endogenous mechanism of gene regulation during development of many eukaryotes. PTGS begins when duplex RNA is recognized within cells by the enzyme Dicer. Dicer is an endonuclease active against dsRNAs. It produces short duplex RNA fragments from 21 to 25 nucleotides long, termed small interfering RNAs (siRNAs) or microRNAs (miRNAs). While siRNAs generally arise from virally produced dsRNAs, miRNAs are transcribed from introns of normal genes or from other nonprotein coding genes, all of which show precise regulation and expression. These miRNA sequences form hairpin structures that are recognized by Dicer to liberate the multiple small miRNAs.
miRNAs interact with a multiprotein complex, termed the RNA-induced silencing complex, RISC. The RISC is composed of a number of components, including the enzyme Argonaut. Argonaut binds the small dsRNA fragments, unwinds them, and incorporates one of the strands (the guide strand) to act as an RNA targeting cofactor; the other strand is degraded. Using the incorporated si/miRNA, the RISC complex can identify sequences complementary to its RNA, whereupon it will either base pair and cleave the complementary RNAs triggering the rapid degradation of complementary RNAs (siRNAs) or will recruit additional proteins that repress the translation of the complementary RNAs via multiple mechanisms (miRNAs).
As many as 1000 miRNAs may function in humans. Some miRNAs target multiple genes; some genes have are targets of multiple miRNAs; and miRNAs are probably involved in some way in regulation of most metabolic pathways. miRNAs regulate central metabolic processes such as adipocyte differentiation and insulin secretion, which are involved in development of obesity and diabetes. Recently, the miR-21 miRNA was identified as an antiapoptotic signal. This miRNA maintains normal apoptotic responses in cells. When miR-21 is aberrantly expressed, as it is in many forms of cancer, it inhibits apoptosis and promotes cellular proliferation.
Summary
 The major products of transcription are the rRNAs, tRNAs, and mRNAs. These RNAs perform specific functions within a cell: mRNAs carry the genetic information from nuclear DNA to ribosomes for protein synthesis; rRNAs interact with proteins to form ribosomes, the basic cellular machinery on which protein synthesis occurs; and tRNAs function as amino acid carriers that translate the information stored in the mRNA nucleotide sequence to the amino acid sequence of proteins.
The major products of transcription are the rRNAs, tRNAs, and mRNAs. These RNAs perform specific functions within a cell: mRNAs carry the genetic information from nuclear DNA to ribosomes for protein synthesis; rRNAs interact with proteins to form ribosomes, the basic cellular machinery on which protein synthesis occurs; and tRNAs function as amino acid carriers that translate the information stored in the mRNA nucleotide sequence to the amino acid sequence of proteins.
 In eukaryotic cells, each of these RNA classes is produced by a different, specific RNA polymerase (RNAPol I, II or III, respectively), while in bacterial cells a single RNA polymerase synthesizes all three classes of RNA.
In eukaryotic cells, each of these RNA classes is produced by a different, specific RNA polymerase (RNAPol I, II or III, respectively), while in bacterial cells a single RNA polymerase synthesizes all three classes of RNA.
 The basic structures of rRNAs and tRNAs in eukaryotic and bacterial cells are similar. However, mRNAs from eukaryotic cells have a 5′ (m7Gppp) cap and a 3′ ([A]n) tail. Prokaryotic mRNA transcripts do not have these modifications on their 5′ and 3′ ends and can be polycistronic.
The basic structures of rRNAs and tRNAs in eukaryotic and bacterial cells are similar. However, mRNAs from eukaryotic cells have a 5′ (m7Gppp) cap and a 3′ ([A]n) tail. Prokaryotic mRNA transcripts do not have these modifications on their 5′ and 3′ ends and can be polycistronic.
 Most, but not all, eukaryotic mRNAs undergo a process called splicing to be functional, whereas prokaryotic mRNAs are functional as soon as they are synthesized. Splicing involves the removal of sequences called introns and the rejoining of exon sequences to each other to form a mature functional mRNA.
Most, but not all, eukaryotic mRNAs undergo a process called splicing to be functional, whereas prokaryotic mRNAs are functional as soon as they are synthesized. Splicing involves the removal of sequences called introns and the rejoining of exon sequences to each other to form a mature functional mRNA.
 The process of transcription consists of three parts: initiation, elongation, and termination. Initiation involves the recognition and binding of promoter sequences by RNA polymerase and associated transcriptional cofactors. Elongation involves the selection of the appropriate nucleotide and formation of the phosphodiester bridges between each nucleotide in an RNA molecule. Finally, termination involves the dissociation of the RNA polymerase from the DNA template. This is mediated by either RNA secondary structure or specific protein factors.
The process of transcription consists of three parts: initiation, elongation, and termination. Initiation involves the recognition and binding of promoter sequences by RNA polymerase and associated transcriptional cofactors. Elongation involves the selection of the appropriate nucleotide and formation of the phosphodiester bridges between each nucleotide in an RNA molecule. Finally, termination involves the dissociation of the RNA polymerase from the DNA template. This is mediated by either RNA secondary structure or specific protein factors.
 Unique cellular mechanisms recognize double-stranded RNA that limit viral infection by multiple mechanisms, some that induce overall mRNA degradation and others that target specific mRNAs for degradation.
Unique cellular mechanisms recognize double-stranded RNA that limit viral infection by multiple mechanisms, some that induce overall mRNA degradation and others that target specific mRNAs for degradation.
Atasoy, D, Betley, JN, Su, HH, et al. Deconstruction of a neural circuit for hunger. Nature. 2012; 488:172–177.
Bora, RS, Gupta, D, Mukkur, TK, et al. RNA interference therapeutics for cancer: challenges and opportunities (review). Mol Med Report. 2012; 6:9–15.
Bumcrot, D, Manoharan, M, Koteliansky, V, et al. RNAi-therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol. 2006; 2:711–719.
Butler, MG. Prader–Willi syndrome: obesity due to genomic imprinting. Curr Genomics. 2011; 12:204–215.
David, CJ, Chen, M, Assanah, M, et al. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010; 463:364–368.
Davis, ME, Zuckerman, JE, Choi, CH, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010; 464:1067–1070.
Ishida, M, Moore, GE. The role of imprinted genes in humans. Mol Aspects Med. 2013; 34:826–840.
Kaiser, PK, Symons, RC, Shah, SM, et al. RNAi-based treatment for neovascular age-related macular degeneration by Sirna-027. Am J Ophthalmol. 2010; 150:33–39.
Keaney, J, Campbell, M, Humphries, P. From RNA interference technology to effective therapy: how far have we come and how far to go? Ther Deliv. 2011; 2:1395–1406.
Liu, H, He, L, Tang, L. Alternative splicing regulation and cell lineage differentiation. Curr Stem Cell Res Ther. 2012; 7:400–406.
Mastroyiannopoulos, NP, Uney, JB, Leonidas, A, et al. The application of ribozymes and DNAzymes in muscle and brain. Molecules. 2010; 15:5460–5472.
Matlin, AJ, Clark, F, Smith, CWJ. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005; 6:386–398.
Zhou, J, Rossi, JJ. Progress in RNAi-based antiviral therapeutics. Methods Mol Biol. 2011; 721:67–75.
www.rcsb.org/pdb/101/motm.do?momID=40
en.wikipedia.org/wiki/RNA_polymeraseα-Amanitin
toxnet.nlm.nih.gov/cgi-bin/sis/search/a?dbs+hsdb:@term+@DOCNO+3458
emedicine.medscape.com/article/1008902-overview
nobelprize.org/nobel_prizes/medicine/laureates/2006/index.html
www.youtube.com/watch?v=cK-OGB1_ELE
www.pbs.org/wgbh/nova/body/rnai-cure.html
www.clinicaltrials.gov/ct2/show/NCT00470977?term=NCT00470977&rank=1
neuromuscular.wustl.edu/pathol/spliceosome.htm
www.rcsb.org/pdb/101/motm.do?momID=65
en.wikibooks.org/wiki/Proteomics/Alternative_splicing_and_its_impact_on_protein_identification
www.eurasnet.info/alternative-splicing/what-is-alternative-splicing