Regulation of Gene Expression
Basic Mechanisms
Introduction
Despite identical DNA is all cells, gene expressions varies significantly with sex, and in time and place in the body
The study of genes and the mechanism whereby the information they hold is converted into proteins and enzymes, hormones, and intracellular signaling molecules is the realm of molecular biology. Except for the erythrocyte, all cells in the body have the same DNA complement. One of the most fascinating aspects of this work is the study of the mechanisms that control differential gene expression, both in time and in place, and the consequences if these control mechanisms are disrupted.
The goal of this chapter is to introduce the basic concepts involved in regulation of protein-coding genes and how these processes are involved in the causation of human disease. The basic mechanism of gene regulation will be described first, followed by a discussion of a specific gene regulation system to highlight various aspects of the basic mechanism. The chapter will end with a discussion of various ways in which the gene regulatory apparatus can be adapted to suit the needs of different tissues and situations.
Basic mechanisms of gene expression
Gene expression is regulated at several different levels
The control of gene expression in humans occurs principally at the level of transcription, the synthesis of mRNA. However, transcription is just the first step in the conversion of the genetic information encoded by a gene into the final processed gene product, and it has become increasingly clear that post-transcriptional events allow for exquisite control of gene expression. The sequence of events involved in the ultimate expression of a particular gene may be summarized as:
At each of these steps conditions allow for the cell to either proceed to the next step or attenuate or halt the process. For instance, if the processing of the RNA is not correct or complete, the resulting mRNA would be useless or possibly destroyed. In addition, if the mRNA is not transported out of the nucleus, it will not be translated. Clearly, during the growth of a human embryo from a single fertilized ovum to a newborn infant, there must be numerous changes in the regulation of genes, to allow the differentiation of a single cell into many types of cells that develop tissue-specific characteristics. Similarly, at puberty there are changes in the secretion of pituitary hormones that result in the cyclic secretion of ovarian and adrenal hormones in females and the production of secondary sexual characteristics. Such programmed events are common in all cellular organisms and the production of these phenotypic changes in cells – and thus the whole organism – arises as a result of changes in the expression of key genes. The expression of genes essential for these processes varies depending on the cell type and the stage of development, but the mechanisms underlying the changes are available to basically all cells. In humans and most other eukaryotes, mechanisms that regulate gene expression are numerous; some of the requirements and options available at each stage are outlined in Table 35.1.
Table 35.1
Requirements and options in the control of gene expression
| Process | Requirements | Options |
| Transcription of mRNA | Chromatin is relaxed (condensed chromatin is a poor template) | Allele-specific transcription |
| DNA in hypomethylated state (methylation of promoter inhibits transcription) | Selection of alternative promoters giving different start sites | |
| Correct trans-acting factors are present (such as transcription factors and cofactors) | ||
| Processing of mRNA | mRNA is 5′ capped | |
| PolyA is added to 3′ end of most messages | Many transcripts are alternatively spliced, increasing coding potential | |
| For most mRNAs the transcript is spliced | mRNAs can be edited to change the coding sequence, changing an amino acid or creating a stop codon | |
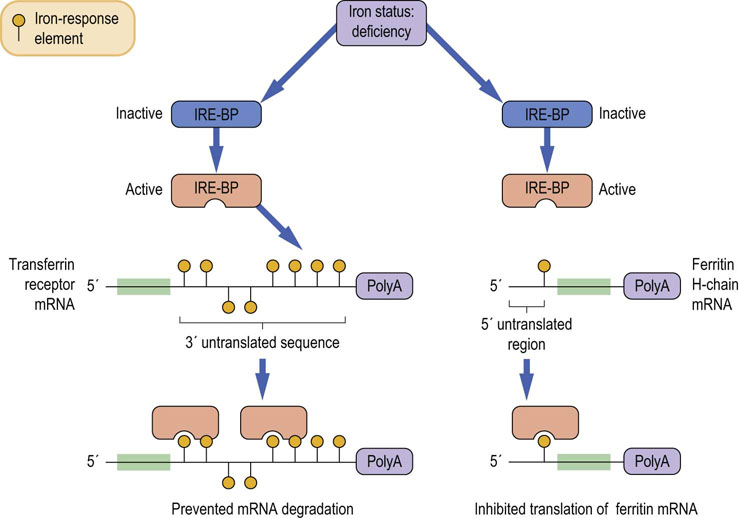
| Signals in the 3′ untranslated regions (UTR) of mRNAs can stabilize or mark the RNA for destruction | ||
| Translation of mRNA | mRNA must be transported to cytoplasm | mRNA can be localized in specific regions of the cytoplasm, such as the ends of axons, for local translation |
| All the factors needed for protein synthesis | Alternative start codons due to internal ribosome entry site (IRES) | |
| Translation on free ribosomes or rough endoplasmic reticulum | ||
| miRNAs can inhibit translation | ||
| Turnover of proteins | Unique protein half-life | Structural proteins tend to turn over slowly collagens |
| Cell cycle proteins are quickly turned over to limit mitosis | ||
| Some proteins contains sequences that target them for rapid degradation |
Gene transcription depends on key elements in the region of the gene
The key step in the transcription of a protein-coding gene is the conversion of the information held within the DNA of the gene into messenger RNA, which can then be used as a template for synthesis of the protein product of the gene. For expression of a gene to take place, the enzyme that catalyzes the formation of mRNA, RNA polymerase II (RNAPol II), must be able to recognize the so-called start points for transcription of the gene. RNAPol II uses one strand of the DNA template to create a new, complementary RNA start point (often called the primary transcript or pre-mRNA), which is then modified in various ways, (commonly including the addition of a 7-methylguanosine cap (m7Gppp) cap at the 5′ end and the polyA tail at the 3′ end, and the removal of introns to form a mature mRNA (Chapter 33). However, RNAPol II cannot initiate transcription alone; it requires other factors to assist in the recognition of critical gene sequences and other proteins to be bound in the vicinity of the start point for transcription.
Promoters
Promoters are usually upstream of the transcription start point of a gene
Sequences that are relatively close to the start of transcription of a gene and control its expression are collectively known as the promoter. Since this is usually within a few hundred or a few thousand nucleotides of the start point, it is usually referred to as the proximal promoter. The promoter sequence acts as a basic recognition unit, signaling that there is a gene that can be transcribed and providing the information needed for the RNAPol II to recognize the gene and to correctly initiate RNA synthesis, both at the right place and using the correct strand of DNA as template. The promoter also plays an important role in determining that the RNA is synthesized at the right time in the right cell. Most control regions in the promoter are upstream (5′) of the transcription start point, and therefore are not transcribed into RNA. Occasionally, some elements of the promoter may be downstream of the start point for RNA synthesis, and may actually be transcribed into RNA. The structure of promoters varies from gene to gene but there are a number of key sequence elements that can be identified within the promoter. These elements may be present in different combinations, some elements being present in one gene and absent in another. Sequences further away from the transcription start site, some of which are known as enhancers, may also have a major impact on the transcription of a gene. These elements are often said to be part of the distal promoter for the gene.
The efficiency and specificity of gene expression are conferred by cis-acting elements
A promoter exerts its effect because it is on the same piece of DNA as the gene being transcribed and is referred to as a cis-acting sequence or element to emphasize that it affects only the neighboring gene on the same chromosome. Since the promoter is critical to gene expression, it is often regarded as being part of the gene it controls, since without it the mRNA would not be made.
The nucleotide sequence immediately surrounding the start of transcription of a gene varies from gene to gene. However, the first nucleotide in the mRNA transcript tends to be adenosine, usually followed by a pyrimidine-rich sequence, termed the initiator (Inr). In general, it has the nucleotide sequence Py2CAPy5 (Py-pyrimidine base) and is found between positions –3 to +5 in relation to the starting point. In addition to Inr, most promoters possess a sequence known as the TATA box approximately 25 bp upstream from the start of transcription. The TATA box has an 8 bp consensus sequence that usually consists entirely of adenine-thymine (A-T) base pairs, although very rarely a guanine-cytosine (G-C) pair may be present. This sequence appears to be very important in the process of transcription, as nucleotide substitutions that disrupt the TATA box result in a marked reduction in the efficiency of transcription. The positions of Inr and the TATA box relative to the start are relatively fixed (Fig. 35.1). Having said this, it must be pointed out that there are many eukaryotic genes that do not have an identifiable TATA box and other sequences are essential for delineating the start of transcription.
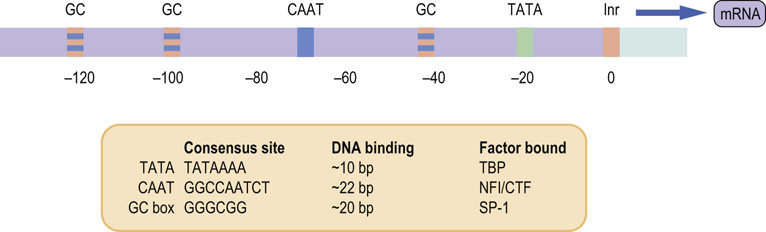
Fig. 35.1 Idealized version of a promoter comprising various elements.
Each promoter element has a specific consensus sequence that binds ubiquitous transcription-activating factors. Binding of transcription factors encompasses the consensus site and a variable number of anonymous adjacent nucleotides, depending on the promoter element. CTF, a member of a protein family whose members act as transcription factors; TBP, TATA-binding protein; NFI, nuclear factor I; SP-1, ubiquitous transcription factor.
In addition to the TATA box, other commonly found cis-acting promoter elements have been described. For example, the CAAT box is often found upstream of the TATA box, typically about 80 bp from the start of transcription. As in the case of the TATA box, it may be more important for its ability to increase the strength of the promoter signal rather than in controlling tissue- or time-specific expression of the gene. Another commonly noted promoter element is the GC box, a GC-rich sequence; multiple copies may be found in a single promoter region.
Figure 35.1 lists some of the common cis-acting elements seen within promoters. These promoter elements bind protein factors (transcription factors) that recognize the DNA sequence of each particular element. Some transcription factors stimulate transcription, others suppress it; some are expressed ubiquitously, others are expressed in a tissue- or time-specific fashion. Thus the array of factors bound to a promoter region can vary from cell to cell, tissue to tissue, and be affected by the state of the organism. These factors, bound to promoter sequences, determine how actively the RNApol II copies the DNA into RNA.
Alternative promoters
Alternative promoters permit tissue or developmental stage-specific gene expression
Although it is clear that a promoter is essential for gene expression to occur, a single promoter may not possess the tissue specificity or developmental stage specificity to allow it to direct expression of a gene at every correct time and place. Some genes have evolved a series of promoters that confer tissue-specific expression. In addition to the use of different promoters that are physically separated, each of the alternative promoters is often associated with its own first exon and, as a result, each mRNA and subsequent protein has a tissue-specific 5′ end and amino acid sequence. A good example of the use of alternative promoters in humans is the gene for dystrophin, the muscle protein that is deficient in Duchenne muscular dystrophy (see Chapter 20). This gene uses alternative promoters that give rise to brain-, muscle-, and retinal-specific proteins, all with differing N-terminal amino acid sequences.
Enhancers
Enhancers modulate the strength of gene expression in a cell
Although the promoter is essential for initiation of transcription, it is not necessarily alone in influencing the strength of transcription of a particular gene. Another group of elements, known as enhancers, can regulate the level of transcription of a gene but, unlike promoters, their position may vary widely with respect to the start point of transcription and their orientation has no effect on their efficiency. Enhancers may lie upstream or downstream of a promoter and may be important in conferring tissue-specific transcription. For instance, a nonspecific promoter may initiate transcription only in the presence of a tissue-specific enhancer. Alternatively, a tissue-specific promoter may initiate transcription but with a greatly increased efficiency in the presence of a nearby enhancer that is not tissue specific. In some genes, for example immunoglobulin genes, enhancers may actually be present downstream of the start point of transcription, within an intron of the gene being actively transcribed.
Response elements
Response elements are binding sites for transcription factors and coordinately regulate expression of multiple genes, e.g. in response to hormonal or environmental stimuli
Response elements are nucleotide sequences that allow specific stimuli, such as steroid hormones (steroid response element; SRE), cyclic AMP (cyclic AMP response element; CRE), or insulin-like growth factor-1 (IGF-1, insulin response element; IRE), to stimulate or repress gene expression. Response elements are often part of promoters or enhancers where they function as binding sites for particular transcription factors. Response elements in promoters are cis-acting sequences that are typically 6–12 bases in length. A single gene may possess a number of different response elements, possibly having transcription stimulated by one stimulus and inhibited by another. Multiple genes may possess the same response element, and this facilitates coinduction or corepression of groups of genes, such as in response to a hormonal stimulus.
Transcription factors
Transcription factors are DNA binding proteins that regulate gene expression
Promoters, enhancers and response elements are part of the gene; transcription factors are the proteins that recognize these structures. These sequence-specific DNA-binding proteins bind to specific nucleotide sequences and bring about differential expression of the gene during development and also within tissues of the mature organism (Fig. 35.2). Many transcription factors act positively and promote transcription, while others act negatively and promote gene silencing. The unique pattern of transcription factors present in the cell will determine in large part which portion of the genome is transcribed into RNA in any given cell. Transcription factors are sometimes referred to as trans-acting factors to emphasize that, as soluble proteins, they can diffuse within the nucleus and act on multiple different genes on different chromosomes.
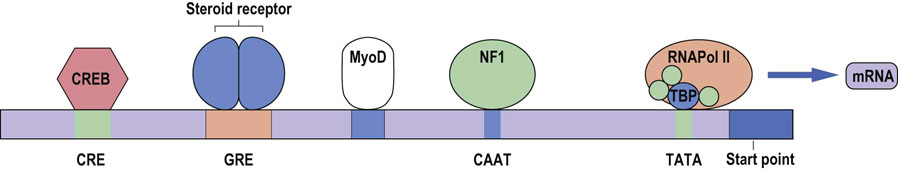
Fig. 35.2 Regulation of gene expression by specific regulatory elements.
Binding of transcription factors to a steroid response element modulates the rate of transcription of the message. Different elements have varying effects on the level of transcription, some exerting greater effects than others, and may also activate tissue-specific expression. CRE, cyclic AMP response element; CREB, CRE-binding protein; GRE, glucocorticoid response element; MyoD, muscle cell-specific transcription factor; NF1, nuclear factor 1. The proteins are shown in a linear array for convenience, but they interact physically with one another, both because of their size and the folding of DNA.
There are other kinds of proteins involved in regulating transcription besides the sequence-specific transcription factors. The so-called general transcription factors, such as TFIIA, B, D, E, etc., form a complex with RNAPol II and this complex is necessary for the initiation of transcription. These general transcription factors are needed for the successful use of every promoter; they vary somewhat with the class of gene, being generally different for RNA polymerase I, II, and III. In eukaryotic cells, and mammalian cells in particular, the RNA polymerases cannot recognize promoter sequences themselves. It is the task of the gene-specific factors to create a local environment that can successfully attract the general factors, which in turn attract the polymerase itself. However, there is emerging evidence that the RNA polymerase complex itself may also be important in the regulation of gene expression.
In addition, other proteins can bind to the sequence-specific transcription factors and modulate their function by repressing or activating gene expression; these factors are often called coactivators or corepressors. Thus, the overall rate of RNA transcription from a gene is the result of the complex interplay of a multitude of transcription factors, coactivators and corepressors. Since there are thousands of these factors in a cell, there is an almost unimaginably large number of combinations that can occur, and thus the control of gene expression can be very specific and very subtle.
In prokaryotes, the cis-elements that control the start site and, in general, the initiation of transcription, are placed closer to the starting point. These cis-elements are fewer in number and there is much less variety (Chapter 33), when compared with those from eukaryotes. In addition, there are fewer trans-acting factors and the control of gene expression is much less subtle in prokaryotes. However, understanding the limitations of the control of gene expression in prokaryotes allows one to appreciate the flexibility of the strategies for control found in eukaryotes.
Initiation of transcription requires binding of transcription factors to DNA
For transcription to occur, transcription factors must bind to DNA. The protein known as TATA-binding protein (TBP) binds to the region of the TATA box. TBP is a general transcription factor, associated with the complex of RNAPol II and a variable number of other proteins. Binding of TBP to the TATA box directs the positioning of the transcription apparatus at a fixed distance from the start point of transcription and thus allows RNAPol II to be positioned exactly at the site of initiation of transcription. Once RNAPol II and a number of other transcription factors have bound to the region of the start point, transcription can occur. When transcription begins, many of the transcription factors required for binding and alignment of RNAPol II are released, and the polymerase travels along the DNA, forming the pre-mRNA transcript.
Transcription factors have highly conserved DNA binding sites
The binding of transcription factors to DNA involves a relatively small area of the transcription factor protein, which comes into close contact with the major and/or minor groove of the DNA double helix to be transcribed. The regions of these proteins that contact the DNA are called DNA-binding domains or motifs, and are highly conserved between species. There are a variety of DNA-binding domains, some of which occur in multiple transcription factors or multiple times in the same factor. Four common classes of DNA-binding domain are the helix-turn-helix and helix-loop-helix motifs, zinc fingers (below), and leucine zippers. Most known sequence-specific transcription factors contain at least one of these DNA-binding motifs and proteins with unknown function that contain any of these motifs are likely to be transcription factors. The average transcription factor has 20 or more sites of contact with DNA, which amplifies the strength and specificity of the interaction.
In addition to a DNA-binding domain, sequence-specific transcription factors also have a transcription-regulatory domain that is required for their ability to modulate transcription. This domain may function in a variety of ways. It may interact directly with the RNA polymerase–general transcription factor complex, it may have indirect effects via coactivators or corepressor proteins, or it may be involved in remodeling the chromatin (below) and so alter the ability of the promoter to recruit other transcription factors.
One way to characterize the interaction of a transcription factor with a particular DNA sequence is to use a technique termed the electrophoretic mobility shift assay (EMSA); (see Fig. 35.3). This method has been used to aid in the purification of transcription factors, to identify them in complex mixtures (such as a cellular extract), to delineate the size of the binding site, and to estimate the strength of the interaction between the factor and the DNA sequence it recognizes.
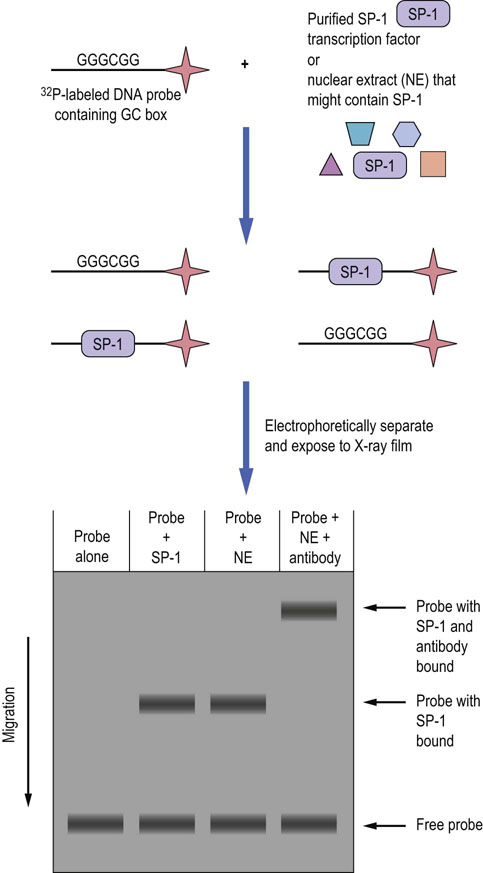
Fig. 35.3 Electrophoretic mobility shift assay (EMSA).
The basic components of the EMSA method are included in the diagram. In this example, a DNA probe that is labeled with 32P is first incubated with either the purified transcription factor SP-1 or with nuclear extract (NE) that contains SP-1, and then subjected to native (nondenaturing) gel electrophoresis. If SP-1 binds to the probe it has a slower mobility in the gel than probe without protein bound (free probe). If an antibody to SP-1 is included in the reaction with NE, then the probe/anti-SP-1 antibody complex migrates even more slowly, confirming that the protein bound to the probe is indeed SP-1. EMSA can be used to help characterize any nucleic acid–protein interaction, including the interaction of RNA and proteins.
Steroid receptors
Steroid receptors possess many characteristics of transcription factors and provide a model for the role of zinc finger proteins in DNA binding
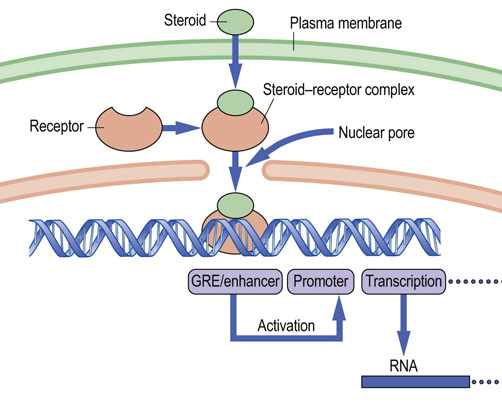
Steroid hormones have a broad range of functions in humans and are essential to normal life. They are derived from a common precursor, cholesterol, and thus share a similar structural backbone (Chapter 17). However, differences in hydroxylation of certain carbon atoms and aromatization of the steroid A-ring give rise to marked differences in biological effect. Steroids bring about their biological effects by binding to steroid-specific hormone receptors; these receptors are found in the cell cytoplasm and nucleus. For the type I (cytoplasmic) receptors, the steroid ligand induces structural changes that lead to dimerization of the receptor and exposure of a nuclear localization signal (NLS); this signal, as well as dimerization, is commonly blocked by a heat shock protein that is released on steroid binding. The ligand–receptor complex now enters the nucleus, where it binds to DNA at a specific response element, the SRE, alternatively called the hormone response element (HRE). SREs may be found many kilobases upstream or downstream of the start of transcription. The steroid–receptor complex functions as a sequence-specific transcription factor and binding of the complex to the SRE results in activation of the promoter and initiation of transcription (Fig. 35.4) or in some cases in the repression of transcription. As might be expected, because of the large number of steroids found in humans, there are correspondingly large numbers of distinct steroid receptor proteins, and each of these recognizes a consensus sequence, an SRE, in the region of a promoter.
The zinc finger motif
A zinc finger motif in steroid receptors binds to the steroid response element in DNA
Central to the recognition of the SRE in the DNA, and to the binding of the receptor to it, is the presence of the so-called zinc finger region in the DNA-binding domain of the receptor molecule. Zinc fingers consist of a peptide loop with a zinc atom at the core of the loop. In the typical zinc finger, the loop comprises two cysteine and two histidine residues in highly conserved positions relative to each other, separated by a fixed number of intervening amino acids; the Cys and His residues are coordinated to the zinc ion. The zinc finger mediates the interaction between the steroid receptor molecule and the SRE in the major groove of the DNA double helix, thus enhancing the efficiency of, and conferring specificity on, the promoter. Zinc finger motifs are generally organized as a series of tandem repeat fingers; the number of repeats varies in different transcription factors. The precise structure of the steroid receptor zinc finger differs from the consensus sequence; the two are compared in Figure 35.5.
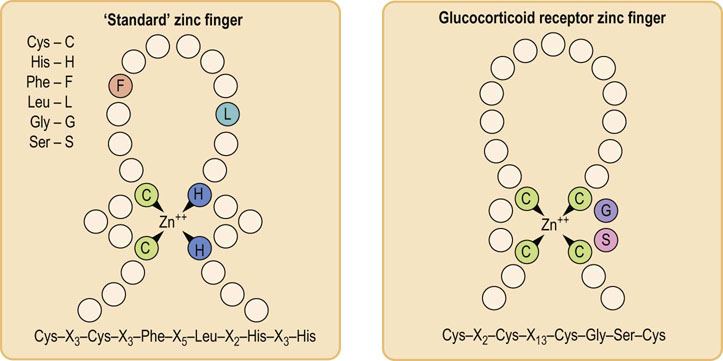
Fig. 35.5 A ‘standard’ zinc finger and a steroid receptor zinc finger.
Zinc fingers are commonly occurring sequences that allow protein binding to double-stranded DNA. C, cysteine; G, glycine; S, serine; X, any intervening amino acid.
Zinc finger proteins recognize and bind to short palindromic sequences of DNA. Palindromes are DNA sequences that read the same (5′ to 3′) on the antiparallel strands, e.g. 5′-GGATCC-3′, which reads the same 5′ to 3′ sequence on the complementary strand. The dimerization of the receptor and recognition of identical sequences on opposite strands strengthen the interaction between receptor and DNA and thus enhance the specificity of SRE recognition.
Organization of the steroid receptor
Steroid receptors are products of a highly conserved gene family
One central feature of all the steroid receptor proteins is the similarity in organization of their receptor molecules. Each receptor has a DNA-binding domain, a transcription-activating domain, a steroid hormone-binding domain, and a dimerization domain. There are three striking features about the structure of the steroid hormone receptors:
 The DNA-binding region always contains a highly conserved zinc finger region, which, if mutated, results in loss of function of the receptor.
The DNA-binding region always contains a highly conserved zinc finger region, which, if mutated, results in loss of function of the receptor.
 The DNA-binding regions of all the steroid hormone receptors have a high degree of homology to one another.
The DNA-binding regions of all the steroid hormone receptors have a high degree of homology to one another.
 The steroid-binding regions show a high degree of homology to one another.
The steroid-binding regions show a high degree of homology to one another.
These common features have identified the steroid receptor proteins as products of a gene family. It would appear that, during the course of evolution, diversification of organisms has resulted in the need for different steroids with varied biological actions and, consequently, a single ancestral gene has undergone duplication and evolutionary change over millions of years, resulting in a group of related but slightly different receptors (Fig. 35.6).
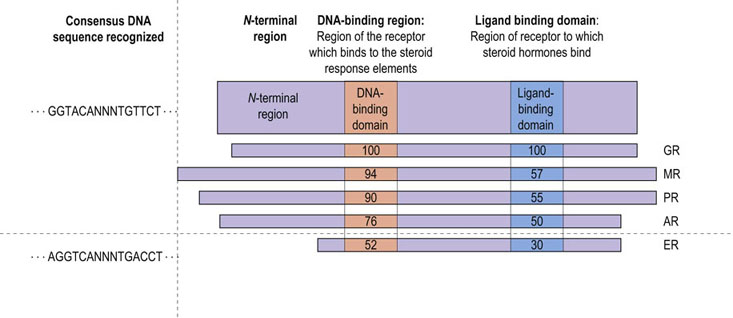
Fig. 35.6 Similarity between different steroid receptors.
The DNA-binding and hormone-binding regions of steroid receptors share a high degree of homology. The estrogen receptor is less similar to the glucocorticoid receptor than are the others. AR, androgen receptor; ER, estrogen receptor; GR, glucocorticoid receptor; MR, mineralocorticoid receptor; PR, progesterone receptor; NNN, any three nucleotides. Numbers denote percent amino acid homology to sequence in GR.
Alternative approaches to gene regulation in humans
Promoter access
Chromatin structure affects access of transcription factors to genes, and thereby affects gene expression
DNA in the cell nucleus is packaged into nucleosomes and higher-order structures in association with histones and other proteins (Chapter 32). Thus, the promoters of some genes may not be readily accessible to transcription factors, even if the transcription factors themselves are present in the nucleus. It has become evident that the degree of packaging of a promoter and the presence, absence or precise location of nucleosomes on a promoter can have major effects on the degree of access for both sequence-specific transcription factors and the RNAPol II complex associated with general transcription factors. Condensed chromatin, termed heterochromatin, where the DNA is tightly associated with the nucleosomes, is usually not a good template for transcription and usually it is necessary for chromatin remodeling (see below) to occur before transcription proceeds. Euchromatin, comprised of DNA that is not as tightly associated with nucleosomes, typically contains regions of active gene transcription. Remodeling may also be necessary in portions of euchromatin, depending on the cell or tissue, but the initial state of the chromatin is less condensed. Portions of a chromosome that are packaged into heterochromatin in one type of cell may be in the form of euchromatin in a different cell, allowing for regulation of gene expression at the level of DNA accessibility. Certain portions of chromosomes, such as the centromeres and telomeres, are examples of regions of the genome that are packaged as heterochromatin in all cells.
Histone packaging, nucleosome stability and therefore the accessibility of DNA are controlled by reversible acetylation and deacetylation of lysine residues in the amino-terminal regions of the core histones, particularly histones H3 and H4. Histone acetyl transferases (HAT) transfer acetyl groups from acetyl-CoA to the amino group of lysines, neutralizing the charge on the lysine residue and weakening the strength of histone–DNA interactions, thereby permitting the relaxation of the nucleosome. Conversely, enzymes that remove the acetyl groups and promote the local condensation of chromosomes are known as histone deacetylases (HDACs).
Reversible acetylation and deacetylation of histones is important in controlling the activation of promoters. Indeed, some transcription factors or transcription coactivators have HAT activity themselves and can remodel chromatin. In the case of some promoters, binding of a transcription factor may result in repositioning of the nucleosomes the next time the DNA replicates, making the gene more likely to be transcribed after cell division. The dynamic interplay of chromatin structure, transcription factor and cofactor binding is important in determining whether a gene is transcribed and how efficiently the RNA polymerase synthesizes it.
Methylation of DNA regulates gene expression
Methylation is one of several epigenetic modifications of DNA; patterns of DNA methylation at birth affect risk for a number of age-related diseases
Certain nucleotides, principally cytidine at the 5 position on the pyrimidine ring, can undergo enzymatic methylation without affecting Watson–Crick pairing. The methylated cytidine residues are usually found associated with a guanosine, as the dinucleotide CpG, and in double-stranded DNA the complementary cytidine is also methylated, giving rise to a palindromic sequence:
The presence of the methylated cytidine can be examined by susceptibility to restriction enzymes (Chapter 36) that cut DNA at sites containing CpG groups only if the CpG is unmethylated, compared to other restriction enzymes that cut whether or not the CpG is methylated. In addition, a bisulfite sequencing technique that relies on the differential reactivity of methyl cytidine can be used to more precisely map the sites of methylation. Many genes in humans (about 50%) have what are called CpG islands (CPI) in the region of their promoters. Generally, these CPI have been found to be unmethylated except in certain pathologic states and cancer. It has become clear that methylation is generally associated with regions of DNA that are less actively transcribing RNA. Demethylation of a promoter may be required for the initiation of transcription, and demethylation of a coding sequence of the gene may also be required for efficient transcription. Regulation of the methylation state of promoters may be a more dynamic process than previously believed, for instance a decrease in the methylation of certain gene promoters in muscle after exercise.
Alternative splicing of mRNA
Alternative splicing yields many variants of a protein from a single pre-mRNA
In Chapter 33, the concept of the splicing of the initial transcript or pre-mRNA was introduced. Most pre-mRNAs can be spliced in alternative ways and the percentage of the alternative splicing of multi-exon transcripts is at least 50%, but with some estimates it may be as high as 80%. This process may provide sufficient diversity to explain individual uniqueness, despite similarities in the gene complement. Thus, by alternative splicing, a particular exon or exons may be spliced out on some but not all occasions. Since most genes have a number of exons (the average is about seven), some pre-mRNAs can eventually give rise to many different versions of the mRNA and, likewise, the final protein. The proteins may differ by only a few amino acids or may have major differences and often have different biological roles. For example, whether an exon is deleted or not may affect where in the cell the protein is localized, whether a protein remains in the cell or is secreted, and whether there are specific isoforms in skeletal versus cardiac muscle. Alternative splicing may also yield a truncated protein, known as a dominant negative mutant protein, that can inhibit the function of the full-length protein. Alternative splicing is regulated, so that particular splice forms may only be seen in certain cells or tissues, at defined stages of development or under differing conditions. In the human brain, there is a family of cell surface adhesion proteins, the neurexins, which mediate the complex network of interactions between approximately 1012 neurons. The neurexins are among the largest human genes, and hundreds, perhaps thousands, of neurexin isoforms are generated from only three genes by alternative promoters and splicing, providing for a diverse range of intercellular communications required for the development of sophisticated neural networks. The neurexins probably have an equally complex set of ligand isoforms, providing tremendous flexibility for reversible cellular interactions during the development of the central nervous system.
Editing of RNA at the post-transcriptional level
The editosome modifies the internal nucleotide sequence of mature mRNAs
RNA editing involves enzyme-mediated alteration of mature mRNAs, before translation. This process, performed by editosomes (Chapter 33) may involve the insertion, deletion or conversion of nucleotides in the RNA molecule. Like alternative splicing, the substitution of one nucleotide for another can result in tissue-specific differences in transcripts. For example, APOB, the gene for human apolipoprotein B (apoB), a component of low-density lipoprotein, encodes a 14.1 kb mRNA transcript in the liver and a 4536-amino acid protein product, apoB100 (Chapter 18). However, in the small intestine, the mRNA is translated into a protein product, called apoB48, which is 2152 amino acids long (~48% of 4536), those amino acids being identical to the first 2152 amino acids of apoB100. The difference in protein size occurs because, in the small intestine, nucleotide 6666 is ‘edited’ by the deamination of a single cytidine residue, converting it to a uridine residue. The resulting change, from a glutamine to a stop codon, causes premature termination, yielding apoB48 in the intestine (Fig. 35.7).
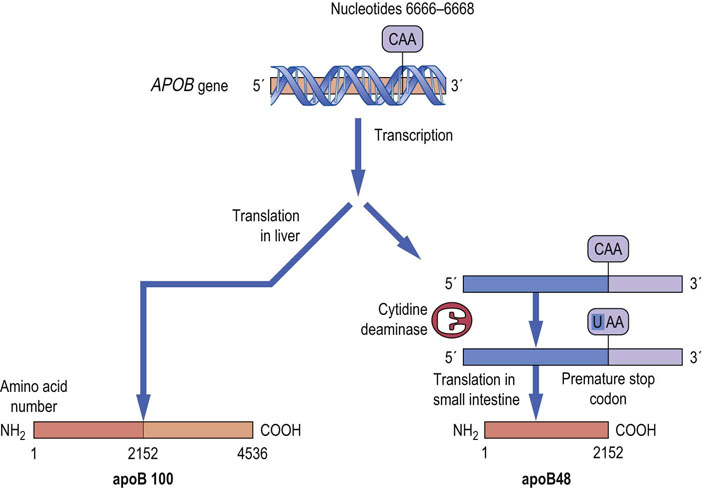
Fig. 35.7 RNA editing of the APOB gene in man gives rise to tissue-specific transcripts.
In the small intestine, nucleotide 6666 of apoB mRNA is converted from a cytosine to uracil by the action of the enzyme cytidine deaminase. This change converts a glutamine codon in apoB100 mRNA to a premature stop codon and, when the mRNA is translated, the truncated product apoB48 is produced. (See also Chapter 18.)
In addition to this cytidine deaminase, there are other enzymes that modify other mRNAs prior to translation, such as the ADARs (adenosine deaminases acting on RNA). ADAR1 catalyzes the deamination of adenosine to inosine residues in dsRNAs; the RNA editing is essential for development of hematopoietic stem cells, and mutations in this enzyme in mice cause early embryonic death. ADAR2 modifies a neuronal glutamate receptor mRNA which results in the change of a single amino acid required for the function of the receptor; deficiency of this enzyme leads to seizures and neonatal death in mice.
RNA interference
RNA interference (RNAi), discussed in more detail in Chapter 33, is another way to control gene expression. At the heart of RNAi are very small noncoding RNAs, about 20–30 nucleotides long, known as micro RNAs (miRNAs). These are involved in the attenuation or repression of translation by binding to the 3′ UTR of an mRNA and recruiting factors that inhibit protein synthesis, or by the destruction of the mRNA by an alternative pathway. During embryogenesis and in certain pathologic states, such as cancer, there are changes in the pattern of miRNA expression in cells, thereby changing gene expression in ways that might alter cell fate or favor cellular proliferation. RNAi holds promise in the treatment of human diseases where the inhibition of the expression of a gene product or the destruction of RNA would be therapeutic, such as in viral infections or cancer.
Preferential activation of one allele of a gene
Human genes are biallelic, but sometimes only one allele of the gene is expressed
The normal complement of human chromosomes comprises 22 pairs of autosomes and two sex chromosomes. In each of the pairs of autosomes the genes are present on both chromosomes: they are biallelic. Under normal circumstances, both genes are expressed without preference being given to either allele of the gene – that is, both the paternal and maternal copies of the gene can be expressed, unless there is a mutation in one allele that prevents this from occurring.
The situation with regard to sex chromosomes is slightly different. Sex chromosomes are of two types, X and Y, the X being substantially larger than the Y. Females have two X chromosomes, whereas males have one X and one Y chromosome. A region of the Y chromosome is identical to a region of the X chromosome but the X chromosome also contains genes that have no matching partner on the Y chromosome, and some genes on the Y chromosome are specific to the Y chromosome, for example SRY, a sex-determining gene. Such genes are said to be monoallelic; they offer no choice as to which allele of the gene will be expressed.
Apart from the specific cases of sex chromosomes, there would appear to be no reason why both alleles of a gene cannot be expressed. However, in humans, genes have been identified that are biallelic but only one allele – either maternal or paternal – is preferentially expressed, despite the fact that both alleles are perfectly normal or identical. As a result, only 50% of the gene product is produced but the product is functionally active. As outlined in Table 35.2, three different mechanisms have been identified that can restrict the expression of biallelic genes in humans (Table 35.2).
Table 35.2
Examples of types of restriction of biallelic genes in humans
| Genomic imprinting | For autosomal genes, imprinting may be tissue specific – monoallelic expression in some tissues, biallelic in others. Examples include insulin-like growth factor 2 (IGF-2) and Wilms' tumor susceptibility gene (WT1) |
| Allelic exclusion | Tissue-specific production of a single allele product; for example, synthesis of a single immunoglobulin light or heavy chain in a B cell from one allele only |
| X chromosome inactivation | Some genes on the X chromosome in females. Males exhibit only one allele of the X-linked gene but females have two, and one is inactivated by switching off nearly the entire X chromosome |
For some genes, although two alleles exist in any particular cell, only one of these alleles is active. Hence the gene behaves as if it were monoallelic although it is, in fact, biallelic.
Summary
 The control of gene expression involves both transcriptional and post-transcriptional events that regulate the expression of a gene in both time and place and in response to numerous developmental, hormonal and stress signals.
The control of gene expression involves both transcriptional and post-transcriptional events that regulate the expression of a gene in both time and place and in response to numerous developmental, hormonal and stress signals.
 DNA sequences and DNA-binding proteins control gene expression. The DNA sequences include cis-acting promoters, such as the TATA box, and enhancers and response elements.
DNA sequences and DNA-binding proteins control gene expression. The DNA sequences include cis-acting promoters, such as the TATA box, and enhancers and response elements.
 The DNA-binding proteins are trans-acting transcription factors that bind with high specificity to these sequences, and facilitate the binding and positioning of RNAPol II for synthesis of pre-mRNA.
The DNA-binding proteins are trans-acting transcription factors that bind with high specificity to these sequences, and facilitate the binding and positioning of RNAPol II for synthesis of pre-mRNA.
 Other factors that affect the conversion of gene to protein include access of the transcriptional apparatus to the gene, enzymatic modification of histones and nucleotides in the DNA, factors that effect alternative intron splicing, post-transcriptional editing of pre-mRNA, RNA interference, and restricted expression of biallelic genes.
Other factors that affect the conversion of gene to protein include access of the transcriptional apparatus to the gene, enzymatic modification of histones and nucleotides in the DNA, factors that effect alternative intron splicing, post-transcriptional editing of pre-mRNA, RNA interference, and restricted expression of biallelic genes.
Barres, R, Yan, J, Egan, B, et al. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metabolism. 2012; 15:405–411.
Berkhout, B, Jeang, KT. RISCy business: microRNAs, pathogenesis, and viruses. J Biol Chem. 2007; 282:26641–26645.
Brena, RM, Costello, JF. Genome-epigenome interactions in cancer. Hum Mol Genet. 2007; 16:R96–R105.
Glaser, KB. HDAC inhibitors: clinical update and mechanism-based potential. Biochem Pharmacol. 2007; 74:659–671.
Goodrich, JA, Tjian, R. Unexpected roles for core promoter factors in cell-type specific transcription and gene regulation. Nat Rev Genet. 2010; 11:549–558.
Kaelin, WG. Use and abuse of RNAi to study mammalian gene function. Science. 2012; 337:421–422.
Iijima, T, Wu, K, Witte, H, et al. SAM68 regulates neuronal activity-dependent alternative splicing of neurexin-1. Cell. 2011; 147:1601–1614.
Li, Q, Lee, J-A, Black, DL. Neuronal regulation of alternative pre-mRNA splicing. Nature Rev Neurosci. 2007; 8:819–831.
Nishikura, K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010; 79:321–349.
Oakley, RH, Cidlowski, JA. Cellular processing of the glucocorticoid receptor gene and protein: new mechanisms for generating tissue-specific actions of glucocorticoids. J Biol Chem. 2011; 286:3177–3184.
Wang, ET, Sandberg, R, Luo, S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008; 456:470–476.
Witt, O, Deubzer, HE, Milde, T, et al. HDAC family: what are the cancer relevant targets? Cancer Letts. 2009; 277:8–21.
Alternate splicing. www.ensembl.org/info/docs/genebuild/ase_annotation.html.
Catalog of genetic diseases. www.ncbi.nlm.nih.gov/omim.
Epigenetics. http://epigenie.com/.
Genomic imprinting. www.geneimprint.com/site/home.
RNA editing. http://dna.kdna.ucla.edu/rna/index.aspx.
RNA interference. www.rnaiweb.com/.