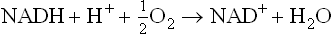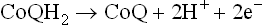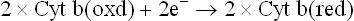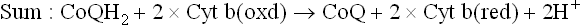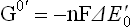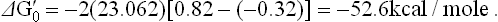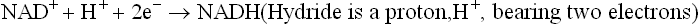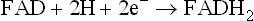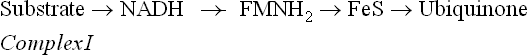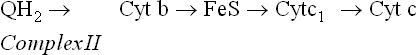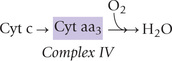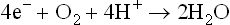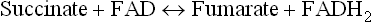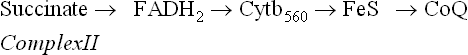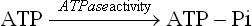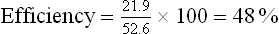Electron Transport, Oxidative Phosphorylation and Mitochondrial Membrane Transporters
Flow of electrons along the electron transport chain (ETC) is the final event in the cellular respiration which releases energy for generation of ATP. The electron transport chains are embedded in the inner mitochondrial membrane. Each chain consists of a series of electron carrying molecules. Electrons move from one carrier protein to another in a stepwise fashion; each carrier being capable of accepting electrons (either with or without accompanying protons) from the preceding carrier and donating them to the next one. At the end of ETC, the electrons reach oxygen, the final acceptor of electrons.
The electrons for the ETC are obtained during catabolism of foodstuffs, e.g. carbohydrates (particularly glucose), fatty acids and amino acids. As these substances undergo oxidation, they lose electrons to the reduction-oxidation cofactors NAD+ and FAD (less commonly to others) to generate NADH and FADH2, respectively. These reduced cofactors can give rise to ATP by transferring their electrons to the electron transport chain. Initially these electrons are energy rich, but as they flow down the ETC, much of their energy is lost. The lost energy is released in small packets, most of which are used for the generation of ATP, while the rest is dissipated (i.e. entropy). Flow of electrons along the ETC (i.e. oxidation) and generation of ATP (i.e. phosphorylation) are coupled processes, together referred to as oxidative phosphorylation. The phosphorylation follows the oxidation; if the latter is inhibited, the former is also similarly affected. Oxidative phosphorylation is described in this chapter followed by a discussion about other carrier proteins that are located in the inner mitochondrial membrane.
At the end of this chapter, the student should be able to understand:
I Electron Transport Chain (ETC)
A Sources of Electrons for ETC
The electrons for the ETC are released during catabolic pathways of biomolecules such as carbohydrates, fats, and amino acids (Fig. 14.1 ) by action of the enzymes known as dehydrogenases. These electrons are then funneled into the ETC. For example, during glycolytic sequence a pair of electrons is removed from glyceraldehyde 3-phosphate. In TCA cycle, an electron pair is removed from each of the following substrates: isocitrate, α-ketoglutarate, succinate and malate by specific dehydrogenases (Table 14.1 ). These dehydrogenases remove a pair of hydrogen atoms initially. Since each hydrogen atom contains an electron, removal of two hydrogen atoms implies removal of an electron pair.
Table 14.1
Action of FAD-linked and NAD+-linked
| Enzyme | Reaction | Location |
| NAD+-linked | ||
| Pyruvate dehydrogenase | Pyruvate → Acetyl CoA | M |
| Isocitrate dehydrogenase | Isocitrate → α-Ketoglutarate | M |
| Malate dehydrogenase | Malate → Oxaloacetate | M |
| α-Ketoglutarate dehydrogenase | α-Ketoglutarate → Succinyl CoA | M |
| Glutamate dehydrogenase | Glutamate → α-Ketoglutarate + Ammonia | M |
| Glyceraldehyde-3-P dehydrogenase | Glyceraldehyde 3-P → 1, 3-Bisphosphoglycerate | C |
| Lactate dehydrogenase | Lactate → Pyruvate | C |
| FAD-linked | ||
| Succinate dehydrogenase | Succinate → Fumarate | M |
| Fatty acyl CoA dehydrogenase | Fatty acyl CoA → Enoyl CoA | M |
M = mitochondrion, C = cytoplasm.
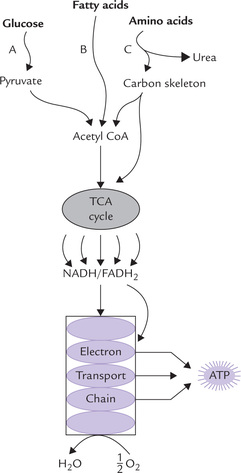
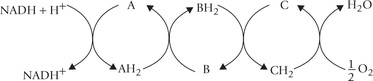
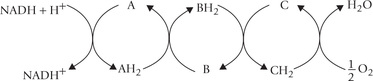
Fig. 14.1 Electron transport and oxidative phosphorylation re-oxidize NADH and FADH2 and trap the energy released as ATP. The NADH and FADH2 arise from all major catabolic pathways: glycolysis, (A); (β-oxidation of fatty acids, (B); amino acid catabolism, (C); and TCA cycle.
These electrons travel down the ETC, and combine with the last acceptor, i.e. oxygen, and two protons are also taken up from the surrounding medium. This results in the formation of water (Fig. 14.2 ).
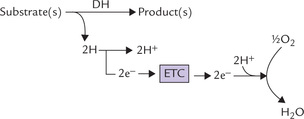
Fig. 14.2 The electron pair, removed from the substrate travels down the electron transport chain (ETC) to the final acceptor, oxygen (DH = dehydrogenase enzyme).
Coenzymes linked with dehydrogenases
The electron pairs removed from the substrate molecules do not enter ETC directly. They are first transferred to specialized coenzymes, nicotinamide adenine dinucleotide (NAD+) or flavin adenine dinucleotide (FAD).
• Certain dehydrogenases, termed NAD+-linked dehydrogenases, transfer the electrons to NAD+-to form NADH.
• Other dehydrogenases, called FAD-linked dehydrogenases, transfer electrons to FAD to form FADH2.
Subsequently the reduced coenzymes, NADH and FADH2, transfer electrons to the ETC at different levels. Some NAD+-linked and FAD-linked dehydrogenases present in catabolic pathways are given in Table 14.1.
B Localization of ETC
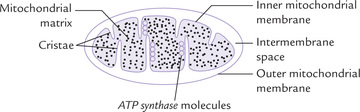
Electron transport chains are located in the inner mitochondrial membrane (IMM). The catabolic pathways that yield electrons for ETC occur in the mitochondrial matrix. Thus, there is close proximity of catabolic pathways with ETC which ensures that the electrons obtained during these pathways are rapidly transferred to ETC (Fig. 14.3 ). Moreover, the ATP synthesizing system is also located in IMM. This permits a rapid utilization of the energy, which was released earlier during the electron flow, for the synthesis of ATP. Since all these processes concerned with oxidative phosphorylation take place in mitochondria, it is appropriate to examine the anatomy of mitochondria in some detail.
Biochemical Anatomy of Mitochondrion
Mitochondrion has two surrounding membranes: the outer mitochondrial membrane and the inner mitochondrial membrane. The outer mitochondrial membrane has a relatively simple structure. It contains special pores which make it permeable to most small molecules and ions. A few enzymes are also located in the membrane. The inner mitochondrial membrane (IMM) has a more complex and specialized structure. It contains a number of protein components such as enzymes, transport proteins, several sets of electron transport chains, and the ATP synthesizing system. A large surface area is required to accommodate all these components. For this purpose, the membrane structure is highly convoluted, being thrown into numerous folds, called cristae, which serve to increase surface area of the IMM several-fold (Fig. 14.3). Thus, the IMM of a single mitochondrion in liver may contain over 10,000 sets of electron transport chains and ATP synthase molecules.
In contrast to the outer mitochondrial membrane which is freely permeable, the inner mitochondrial membrane is selectively permeable, meaning that it is impermeable to most ions (H+, K+, Na+) and small molecules (ADP, ATP), and permeable only to a few: only water, carbon dioxide and oxygen can freely move across the IMM. Movement of other substances across the membrane can take place only through mediation of specific transport proteins.
Presence of these protein components makes the IMM unusually rich in proteins which constitute 75% or more of the total membrane weight. Rest of the membrane structure is formed by lipids. This contrasts with the other cell membranes which contain less amount of proteins (Chapter 7).
Mitochondrial matrix is the space enclosed by the inner mitochondrial membrane. It contains a gel-like solution in which several catabolic pathways, e.g. citric acid cycle, β-oxidation of fatty acids, and oxidation of amino acids, occur. In addition to enzymes of these pathways, the matrix contains coenzymes such as NAD+, NADP+ and FAD, and components of the phosphorylation reaction, e.g. ADP, ATP and phosphate ions.
C Electron Transport: An Overview
ETC consists of several electron carriers that are arranged in a sequence shown in Figure 14.4 .
The electrons are transported from one carrier molecule to the next either as hydride ion (:H-) which bears two electrons (discussed later), or as hydrogen atoms or as free electrons.
The starting compound, that loses its electrons to the ETC is NADH and the final acceptor of the electrons is oxygen. NADH is formed by transfer of energy-rich electron pair removed from a substrate during catabolism, as mentioned earlier (equation i). NADH loses its electrons to the first carrier of ETC (NADH dehydrogenase). Thereafter, the electrons pass down from one carrier to another. During the electron transfer, the components of ETC accept or donate electrons, either with or without accompanying protons. A simple transport chain is depicted as below.
The sum of all these reactions is:
The electrons may travel, not only as free electron (or hydride) but also as hydrogen atoms. This is logical as a hydrogen atom consists of one electron and one proton.
D Redox Couples and Redox Potential
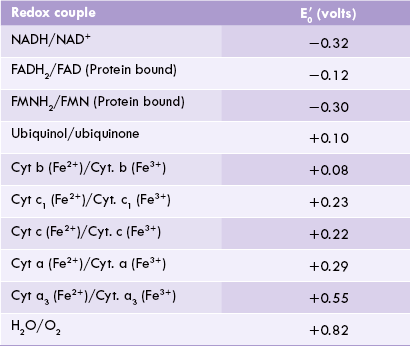
Why do electrons flow unidirectionally from NADH (or FADH2) to oxygen, and what is the thermodynamic origin of the high energy bonds being created? The answer may be obtained by looking at the redox potentials of various carriers. As noted, a given carrier oscillates between the reduced and the oxidized states. For example, ubiquinone (CoQ) interchanges between a reduced form, i.e. ubiquinol or CoQH2 and an oxidized form, i.e. ubiquinone or CoQ. The reduced and the oxidized forms of the same carrier are together referred to as redox couple. Thus, the CoQ and the CoQH2 constitute redox couple, as do the oxidized cytochrome b and the reduced cytochrome b.
Flow of electrons between these two redox couples is shown in an expanded form below:
The tendency of a redox couple to donate or accept electrons is determined by standard redox potential (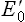 ). Every redox couple has a characteristic value of
). Every redox couple has a characteristic value of 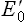 . Electrons always flow from the lower to the higher redox potential, so that redox systems with negative
. Electrons always flow from the lower to the higher redox potential, so that redox systems with negative 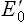 tend to donate electrons to redox systems with positive
tend to donate electrons to redox systems with positive 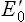 . For example:
. For example:
• Standard redox potential of NADH/NAD+ is -0.32 V.
• That of CoQH2/CoQ is +0.10 V.
• Therefore, direction of flow of electrons is from NADH/NAD+ to ubiquinol/ubiquinone and never the opposite way.
For the same reason, electrons flow from FMNH2/FMN (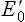 –0.30 V) to CoQH2/CoQ (
–0.30 V) to CoQH2/CoQ (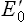 +0.05 V). In this way, all components of the respiratory chain are arranged to range from the most negative
+0.05 V). In this way, all components of the respiratory chain are arranged to range from the most negative 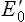 (–0.32 V of NADH/NAD+) to the most positive
(–0.32 V of NADH/NAD+) to the most positive 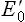 ( + 0.82 V of H2O/O2) pair.
( + 0.82 V of H2O/O2) pair.
Standard redox potential of components of ETC are shown in Table 14.2 .
E Free Energy Changes During Electron Flow
Just as all spontaneous transformations proceed in such a direction that there is a loss of free energy, the electron transfer between the redox pairs is also accompanied by release of free energy. Amount of free energy released is directly proportional to a difference in the standard redox potentials of the redox pairs. A definite relationship exists between the two as below:
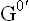 = standard free energy change in kcal/mole.
= standard free energy change in kcal/mole.
n = number of reducing equivalents (i.e. electrons) transferred (its value is 2 in the present case, as the electrons are transferred in pairs).
F = Faraday’s constant (23.062 kcal V-1 mol-1).
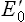 = difference between the standard redox potentials of the electron-donor and the electron-acceptor redox systems.
= difference between the standard redox potentials of the electron-donor and the electron-acceptor redox systems.
The given relationship holds good under standard conditions, i.e. concentration of 1.0 M of both the electron donor and the electron acceptor, temperature 25°C and pH 7.0. It can be used to calculate the standard free energy change as a pair of electrons passes along the electron transport chain, from the first redox couple (i.e. NADH/NAD+; 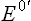 = –0.32 V) to the last one (i.e. H2O)/ O2;
= –0.32 V) to the last one (i.e. H2O)/ O2; 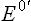 = +0.82 V).
= +0.82 V).
This relationship can also be used to calculate the energy changes for individual segments of the electron transport chain.
F Components of Electron Transport Chain
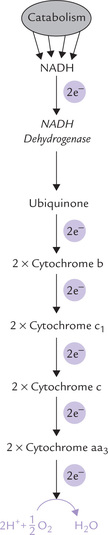
Five distinct types of components are present in ETC: (a) Nicotinamide nucleotides, (b) flavoproteins (containing flavin mononucleotide or FMN; and flavin adenine dinucleotide or FAD), (c) iron-sulphur centres, (d) ubiquinone, and (e) cytochromes. Their arrangement in the respiratory chain has been depicted in Figure 14.5 .
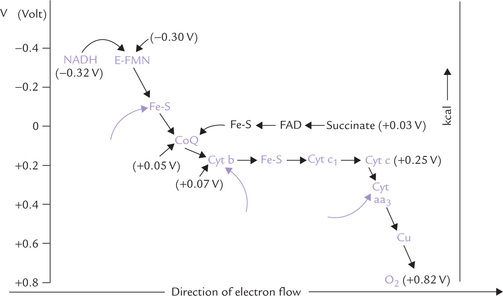
Fig. 14.5 The respiratory chain of mitochondria showing direction of flow of electrons and energy relationships. The arrows (→) indicate the sites of ATP generation (E = NADH dehydrogenase, Fe-S = iron-sulphur centres, CoQ = coenzyme Q or ubiquinone, cyt = cytochrome).
Nicotinamide Nucleotides
Nicotinamide adenine dinucleotide (NAD+) and nicotinamide adenine dinucleotide phosphate (NADP+) are the coenzymes derived from the vitamin niacin. The former is more actively involved in the ETC. It is reduced to NADH by transfer of a pair of electrons (in form of hydride ion) from the substrate by action of various dehydrogenases (see NAD+-linked dehydrogenases; Table 14.1).
NADH subsequently loses its electrons to the initial component of ETC: NADH dehydrogenase, a large protein complex embedded in IMM.
NADP+ is similarly produced by action of NADP-dependent dehydrogenases, but it is mostly involved in reductive biosynthesis of biomolecules.
Flavoproteins
Flavoproteins are important hydrogen carriers. Their prosthetic groups are flavins (FMN or FAD), which are derivatives of the vitamin riboflavin. These coenzymes have chemically related structures. Both have a flavin mononucleotide (FMN) unit, which contains the reactive site. FAD has an additional sugar group and an adenine base, which completes its structure (Chapter 18). FAD and FMN react with two protons plus two electron, in alternating between the reduced and the oxidized state:
FMN is prosthetic group of NADH dehydrogenase. It is bound firmly to the enzyme protein and does not function as a diffusible co-substrate. FAD, on the other hand, is linked to another flavoprotein, succinate dehydrogenase, and functions as a diffusible co-substrate.
Iron-sulphur Centres
They are also known as non-haem iron proteins or the iron-sulphur clusters. Several types of iron sulphur (FeS) centres exist, but in each case the iron atoms are coordinated to inorganic sulphur atoms (and the sulphur of cysteine side chains in proteins). Within an FeS cluster, an electron is carried by the iron atom, which, upon accepting the electron, changes from the Fe+3 (ferric) state to the Fe+2 (ferrous) state. As the electron is passed to another electron carrier, the iron atom of the FeS cluster changes back again to the ferric state.
As explained in the next section, one group of iron-sulphur protein participates in the transfer of electrons from FMN to ubiquinone (CoQ), and the other from cytochrome b to cytochrome c1 (Fig. 14.5).
Ubiquinone
Ubiquinone, also known as coenzyme Q, is a mobile diffusible hydrogen carrier, which can move from donor to acceptor molecules during electron transport. It is a benzoquinone with a hydrocarbon tail of (usually) 10 isoprene (branched 5-carbon) units (Fig. 14.6 ), which makes it strongly hydrophobic and confines it to the lipid bilayer of the inner mitochondrial membrane. It was also named ubiquinone because of its ubiquitous or widespread distribution.
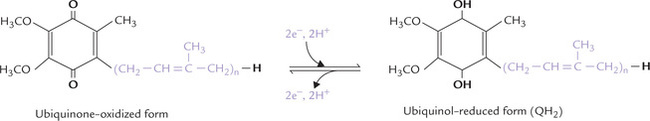
Fig. 14.6 Structure of ubiquinone, also called coenzyme Q (CoQ) or UQ. The isoprene units are shown in colour.
The names coenzyme Q and ubiquinone and the abbreviations CoQ and UQ are used interchangeably. A subscript(n) indicates the number of isoprene units; for example, CoQ6 or CoQ10 contain 6 or 10 isoprene units respectively. CoQ10is the homologue that occurs in humans. It can accept electrons from FMNH2 or from FADH2 and transfers them to cytochromes.
Generally, ubiquinone carries two hydrogen atoms, but can also act in one-electron transfers by forming a free-radical intermediate, called semiquinone intermediate.
The reduced form is called ubiquinol (Fig. 14.6).
Cytochromes
Cytochromes are the final class of components that participate in electron transport. They are integral membrane proteins (with the exception of cytochrome c), and are named so because they are coloured cellular components (cyto cell, chrome colour). They are conjugated proteins, containing an iron-porphyrin as a prosthetic group. In most cases, this iron-porphyrin is the haem group, also found in oxygen transport proteins, e.g. haemoglobin and myoglobin. In contrast to these proteins, however, in cytochromes the haem iron cannot bind oxygen (or carbon monoxide or any other potential ligand), but rather acts as a reversible electron carrier. Iron in cytochromes (but not in haemoglobin or myoglobin) undergoes physiological oxidation-reduction between the ferrous (+2) and ferric (+3) states, as electrons are shuttled from one protein to another.
Why cytochromes can carry only electrons, but cannot bind oxygen? This is shown diagrammatically in Figure 14.7 . In cytochromes, the iron group of the haem is anchored on both sides, on one side by histidine and on the other by methionine. In haemoglobin, it is only anchored on one side by histidine, enabling oxygen to interact with the free side of Fe.
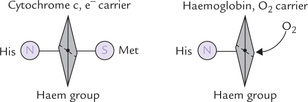
Fig. 14.7 Haem is prosthetic group in both cytochrome C and haemoglobin, but performs different roles. It serves as oxygen carrier in haemoglobin and electron carrier in cytochrome C (His = histidine; Met = methionine).
The main varieties of cytochromes in mitochondria are the a, b, and c types of cytochromes, the classification being based on the respective absorption spectrum and the type of porphyrin structure present. The cytochrome of b type contains haem, whereas the others have porphyrins with different side chains. Cytochromes a and a3 contain “haem a” rather than “ordinary haem”. In this the porphyrin ring of haem is modified: (i) a methyl group of haem is oxidized to a formyl group, and (ii) a hydrophobic isoprenoid chain is attached to one of the vinyl groups. (See chapter 16 for haem structure.)
The cytochrome families are subclassified in terms of the historical order of their discovery (b1, b2, b3...) or the wavelength (nm) of a characteristic spectral absorption peak (b562, b566). The electrons are transported from coenzyme Q to cytochromes in the order: b, c1, c, a and a3. The last two can directly react with molecular oxygen. Besides haem iron, the aa3are associated with copper, which acts as an electron carrier by a valence change between the cuprous (Cu+) and cupric (Cu2+). As described later, it (aa3) participates in the last reaction and transfer of electrons to molecular oxygen.
G Pathway of Mitochondrial Electron Transport
Only four components of the respiratory chain are freely diffusible (NADH, ubiquinone, cytochrome c and oxygen), and the rest are organized as constituents of large protein complexes (Fig. 14.8 ). There are four complexes (I to IV) embedded in the inner mitochondrial membrane. Exact detail of the complexes is not known. What is known is: (i) that a complex contains many polypeptide or protein subunits and several iron centres, (ii) that these components can be readily reduced or oxidized, and (iii) that they transfer electrons as they flip between the reduced state and the oxidized state.
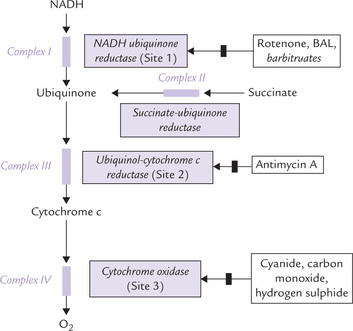
Fig. 14.8 The structured complex of functionally related electron carriers. Inhibitors of ETC ( ) are also shown. Sites 1, 2 and 3 refer to the sites of proton pumping (“phosphorylation sites”).
) are also shown. Sites 1, 2 and 3 refer to the sites of proton pumping (“phosphorylation sites”).
Complex I
Oxidation of NADH begins with Complex I (also called NADH-ubiquinone reductase or NADH dehydrogenase complex), which comprises 28–41 protein subunits (depending upon the species), FMN as a prosthetic group and about 7-iron-sulphur (FeS) centres. It transfers electrons from NADH to ubiquinone via FMN and FeS centre:
As the electron pair flows from NADH to complex I, it is accepted together with a hydrogen ions, H+, such that two electrons and two H+ are accepted in total. As a result FMN is converted into FMNH2. The electrons are then transferred within the complex I to iron-sulphur clusters, and then passed onto ubiquinone, which is thereby converted to ubiquinol (Fig. 14.6). Ubiquinone has a long, flexible, lipid soluble arm, and so it can readily move through the inner membrane to transfer the electrons to the next enzyme in the sequence, i.e. the Complex III (the Complex II will be dealt with later).
Complex III
Complex III (ubiquinol-cytochrome c reductase complex) once again uses iron atoms to shuffle electrons within its structure. It contains cytochrome b, an iron-sulphur centre (called Rieske’s protein), and cytochrome C1.
Figure 14.9 elaborately depicts the electrons passing from ubiquinol, through the cytochrome b, FeS and cytochrome c1 components of this complex, and the accompanying ferric-ferrous interconversions.
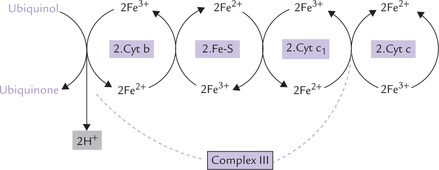
Fig. 14.9 Flow of electrons from ubiquinol to cytochrome c via the large, multiprotein complex III, consisting of cytochrome b, an iron sulphur protein and cytochrome c1.
Note that cytochrome c is a peripheral membrane protein bound to the outer membrane surface, which transfers its electrons to Complex IV.
Complex IV
Complex IV (cytochrome oxidase complex) contains 13 different polypeptides, two haem groups, and two copper ions. The complex IV transfers electrons to O2, the final acceptor, to form water:
The complex contains two cytochromes (a and a3), which are associated with haem iron and copper. Cytochrome a is paired with a copper atom, CuA and cytochrome a3 is paired with a different copper atom, CuB. During electron transfer, the iron atoms of the haem cycle between the ferric and ferrous states while the copper atoms cycle between cuprous and cupric. The cytochrome oxidase reaction is complex: it transfers four electrons to molecular oxygen to form two molecules of water. Oxygen is tightly bound between haem a3 and copper during its reduction, and it is released only after its complete reduction to water.
Because of high affinity of cytochrome oxidase for molecular oxygen, the oxidative phosphorylation is near maximum even at low oxygen pressure.
Complex II
Complex II (succinate-ubiquinone reductase or succinate dehydrogenase) contains four polypeptide chains; the first two constitute the bonafide succinate dehydrogenase (SDH), a Krebs cycle enzyme, which catalyzes the following reaction:
SDH contains iron-sulphur centres, in addition to the covalently bound FAD. Electrons from FADH2 are transferred to ubiquinone (via cytochrome b560 and FeS) and reduce it to ubiquinol. The latter then proceeds to reduce Complex III.
Evidently, succinate dehydrogenase (and other mitochondrial flavoproteins) bypasses the Complex I and pass their electrons directly to ubiquinone. This results in generation of only two ATPs when FADH2 is used as the substrate; three are generated if NADH is used as the substrate. The reason for this is that one ATP each is generated at Complex I, III, and IV.
H Inhibitors of ETC
A number of site-specific inhibitors are known to block transport of electrons along the respiratory chain (Fig. 14.8). This action inhibits oxygen consumption and secondarily halts synthesis of ATP from ADP and Pi. Some common inhibitors and their sites of action are as follows:
Rotenone
It blocks transfer of electrons from NADH to ubiquinone. It is a plant product that is extremely toxic and is used by the American Indians as a fish poison.
However, it is not very toxic to humans because of its poor intestinal absorption. Humans, therefore, can safely eat the poisoned fish.
Barbiturates
Besides rotenone, some barbiturates (e.g. amytal) inhibit electron flow through Complex I.
Antibiotics
An antibiotic, British antilewisite (BAL) also acts at Complex I; and another antibiotic, antimycin A, inhibits Complex III.
Cyanide, carbon monoxide azide, and hydrogen sulphide
These inhibitors act at complex IV. They bind to the iron protoporphyrin in cytochrome aa3 whose sixth coordination position is not occupied by an amino acid side chain but is reserved for oxygen. Cyanide and azide bind to the ferric form of the iron; carbon monoxide binds to the ferrous form.
Hydrogen sulphide also is a potent inhibitor. Carbon monoxide not only inhibits electron flow, but also has high affinity for haemoglobin, which adds to its clinically important toxicity.
Inhibition of the electron flow by these agents creates crossover points. The electron carriers present before the block become reduced and those after the block become oxidized. Since absorption spectra of the oxidized and the reduced forms of electron carriers differ from each other, these changes can be detected by spectrophotometer.
II Oxidative Phosphorylation
There are several hypotheses proposed to explain how energy released during flow of electrons along the respiratory chain is used for ATP generation.
A Hypotheses for ATP Generation
Chemical Coupling Hypothesis
This hypothesis, proposed by Edward Stater (1953), postulates that the energy released from ETC causes formation of high energy covalent intermediates. These intermediates are subsequently cleaved to release their energy content, which is used for the synthesis of ATP.
Conformational Coupling Hypothesis
The conformational coupling hypothesis put forth by Paul Boyer (1964), proposes that the energy of electron transport is used for altering conformation of certain proteins that are located in the IMM. The proteins with altered conformation have high energy content, which is subsequently used for ATP generation.
Chemiosmotic Hypothesis
Proposed by the British biochemist Peter Mitchell (1961), this hypothesis is widely accepted. According to the hypothesis, oxidative phosphorylation occurs in two steps:
• Generation of electrochemical gradient across the IMM (Step I).
• Utilization of this gradient to fuel ATP synthesis (Step II).
Step I
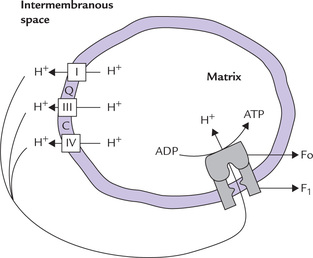
The electrochemical gradient is built by pumping of protons out of the mitochondrial matrix. The proton pumping is fueled by the energy released by exergonic redox reactions of ETC (Fig. 14.10 ). This creates a proton gradient of approximately 1.4 pH units across the IMM (inside alkaline). It also builds a membrane potential of 100–200 mV1 (inside negative). The concentration and electrical potential add up to a steep electrochemical gradient for protons.
Step II
The electrochemical gradient is a potential source of energy (4–6 kcal/mol) for generation of ATP. The enzyme responsible for utilizing this gradient for ATP synthesis is known as ATP synthase. It is also known as the ATPase, because the isolated enzyme is capable of catalyzing hydrolysis of ATP to ADP and inorganic phosphate.
Figure 14.11 gives a magnified view of the ATP synthase, which consists of two components: F0 and F1. The F0 is embedded in the IMM, whereas the F1 protrudes into the mitochondrial matrix (Fig. 14.11). F1 unit (F1 stands for “coupling factor 1”) is a protein complex of subunit structure δ3β3γδ and a molecular weight of 380 dDa. It can be visualized in the electron microscope as small buttons on the inner surface of the inner mitochondrial membrane. The F1 unit is attached to the F0unit, an integral membrane protein having a pore called proton channel.
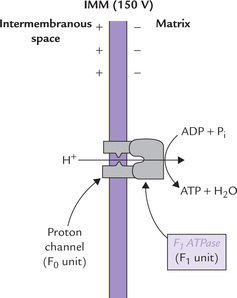
Fig. 14.11 Protons are allowed to flow back into the mitochondrion through a specific proton channel in F0. This proton channel is coupled to an ATP synthesizing enzyme (the F1 unit).
The proton channel is crucial for ATP generation. This is because the IMM is impermeable to protons, and so the extruded protons in the intermembrane space can reenter the mitochondrial matrix through this proton channel. As these protons move inwards, the energy inherent in the electrochemical gradient is liberated as concentrated packets. These energy packets are readily used by the F1-unit to fuel ATP synthesis.
Action of the ATP synthase is inhibited by an antibiotic, oligomycin. The latter binds with F0 portion of this enzyme and closes the proton channel. As a result, reentry of protons is blocked and the energy inherent in the electrochemical gradient cannot be used for ATP generation. As more and more protons are actively pumped out, the proton concentration in the intermembrane space rises. It becomes difficult to pump protons against the rising gradient and finally electron transport also stops. At this stage, the ATP synthase is turned off.
Success of chemiosmotic hypothesis: The chemiosmotic hypothesis has been widely accepted because it can provide explanations for the following phenomena:
1. An intact membrane is required for ATP synthesis by oxidative phosphorylation.
2. Respiring mitochondria generates a proton gradient.
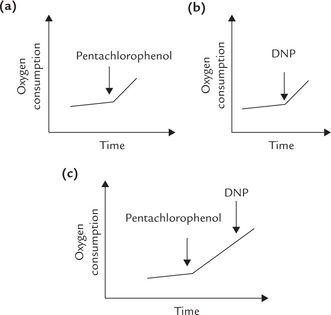
3. Certain compounds (such as DNP) stop ATP synthesis without inhibiting electron transport from NADH, or succinate, to oxygen. These compounds prevent building up of the proton gradient, which stops/ halts synthesis of ATP. Thus, they act as uncouplers of oxidative phosphorylation (discussed later).
B ATP Production and P : O Ratio
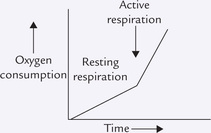
It is well established experimentally that a pair of electrons originating in NADH generates three molecules of ATP via the oxidative phosphorylation pathway, and that from FADH2 generates two. It is said that the P : O ratio is three for electrons from NADH and two for those from FADH2. P referring to a high-energy phosphate bond being synthesized and O referring to an atom of oxygen being reduced. Thus P : O ratio is defined as the number of ATPs generated for each oxygen consumed. Refer to Box 14.1 for more information on relationship between ATP generation and oxygen consumption.
Sites of ATP Production
There are three energy conserving segments that release sufficient amount of energy to result in synthesis of an ATP molecule each. At each of these segments, called phosphorylation sites (indicated by arrows in Fig. 14.5), quantum of energy released exceeds 10 kcal/mole. Because ATP is synthesized only when protons flow, this results in halting ATP synthesis.
1. The Site I is between NADH and ubiquinone (specifically during electron flow through the FeS complex).
2. The Site II is between ubiquinol and cytochrome c (specifically during electron flow from cytochrome b to cytochrome c1).
3. The Site III is between cytochrome c and oxygen (i.e. the cytochrome oxidation reaction).
The three sites correspond to Complexes I, III and IV of the ETC. Since Complex I is bypassed with FADH2, only two ATPs are generated (P : O ratio-2).
C Energetics
If one starts from NADH, with its redox potential of -0.32 V, and ends with oxygen, redox potential +0.82 V, then one spans a total of 1.14 V (Fig. 14.5). As discussed earlier, this comes to a theoretical availability of about 52.6 kcal/mole (equation iv). Because three ATP molecules are synthesized for each pair of electrons channeled to oxygen, oxidative phosphorylation recovers 21.9 kcal (each ATP = 7.3 kcal) of the 52.6 kcal available as ATP, for an approximate efficiency of 48%.
The remaining 52% of energy is lost as heat.
Uncouplers
Being linked through a proton gradient, the oxidation and the phosphorylation are said to be coupled processes. They can be uncoupled from each other by certain compounds, called the uncouplers. Primary action of these compounds is to increase permeability of the inner mitochondrial membrane to protons. As a result, relatively free movement of protons across the IMM occurs, which prevents building of the electrochemical gradient. Since this gradient is essential for ATP generation, failure to build it stops ATP generation.
An example of uncoupler is 2, 4-dinitrophenol (Fig. 14.12 ). The compound can freely move across the IMM due to its lipophilic nature. It can carry a proton along with itself. As a result, free movement of protons across the IMM occurs, preventing the building up of proton gradient. Though an uncoupler impedes ATP generation, it has no effect on electron transport. The latter process continues as before, or is rather enhanced due to acceptor control of respiration, discussed later. The energy released by electron flow is mostly dissipated (i.e. entropy). Some portion of it generates heat: this explains thermogenic effect of some uncouplers (Case 14.1).
Fig. 14.12 Structure of 2,4 dinitrophenol, a highly diffusible chemical that uncouples oxidative phosphorylation by transporting protons across the IMM.
The other uncouplers include pentachlorophenol, dinitrocresol, and trifluorocarbonylcyanide. The latter dissipates the proton gradient 100 times faster than DNP.
Physiological uncouplers and ionophores
Some endogenous compounds, including free fatty acids, bilirubin and possibly thyroxine, also can act as uncouplers at concentrations well above the physiological range. They are called physiological uncouplers. In case of bilirubin, which deposits in the basal ganglia of infants with severe hyperbilirubinaemia (see Chapter 16), a role of the uncoupling effect in the resulting brain damage is possible.
Valinomycin is a transport antibiotic or an ionophore that makes the inner mitochondrial membrane permeable to potassium (the term ionophore refers to compounds that promote the transport of ions across biological membranes; Chapter 7). It also acts as an uncoupler.
This is because the translocation of potassium ions dissipates the membrane potential, which is an essential component of the proton-motive force.
Significance of Uncoupling
Uncoupling under natural conditions plays an important role in hibernating animals, and in animals of polar regions, for maintaining their body temperature. These animals contain brown adipose tissue which is specialized to carry out oxidation uncoupled from phosphorylation. When fats are oxidized in the brown adipose tissue, the energy liberated is not trapped as ATP but rather released as heat.
Presence of active brown adipose in some individuals is believed to burn excess calories, thereby preventing them from gaining weight even after indulging in overeating.
III Mitochondrial Membrane Transporters
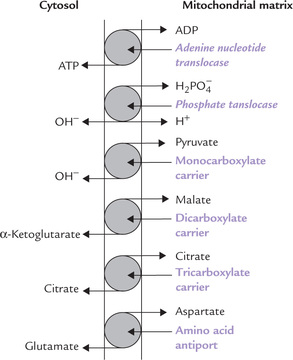
The carrier proteins in IMM, also known as mitochondrial membrane transporters, are highly specific in their action. They permit movements of several molecules that are important for mitochondrial function, such as ATP, H+, pyruvate, malate, citrate, etc. A few transport proteins (Fig. 14.13 ) and their actions are described here.
A Adenine Nucleotide Translocase (ANT)
The adenine nucleotide translocase permits inward movement of ADP into the mitochondrial matrix, and a simultaneous outward movement of ATP into the intermembranous space. These movements are important because ADP (and phosphate) have to enter the mitochondrion as substrates for oxidative phosphorylation, and ATP has to pass from the mitochondrion to the cytosol, where it is used for energy-dependent processes. Structurally, ANT is an integral membrane protein that extends across the IMM.
Atractyloside is toxic glycoside which specifically inhibits the ANT system. Inhibition of ANT hampers the transport of ATP, which leads to serious consequences since ATP is required to drive a number of cellular activities.
B Phosphate Translocase System
The phosphate translocase system is functionally related to the adenine nucleotide translocase. It cotransports a phosphate ion along with H+ into the mitochondrial matrix (Fig. 14.13). Phosphate is then used for the generation of ATP from ADP. Thus, a combined action of these two translocase systems permits entry of phosphate and ADP into the mitochondrial matrix, where they combine to form ATP.
C Monocarboxylate Carrier
This carrier transports pyruvate produced in cytosol, mainly through the glycolytic sequence, into mitochondria where it is further metabolized.
D Dicarboxylate Carrier
It moves malate from site of its production (i.e. the mitochondrial matrix) into cytosol.
E Tricarboxylate Carrier
It transports citrate, the first intermediate of TCA cycle, into cytosol. This carrier is very specific: it does not transport even a closely related molecule, i.e. isocitrate.
Combined action of the dicarboxylate and tricarboxylate carriers play vital role in lipogenesis (Chapter 11).
F Shuttle Systems
Certain shuttle systems are known to operate in mitochondria that transport reducing equivalents from cytosol into mitochondria. The reducing equivalents (in the form of NADH) are generated in the cytosol (during glycolysis). They have to be transported to mitochondrial matrix for oxidation and generation of ATP. However, the IMM is impermeable to NADH. Therefore, specific shuttle systems, namely malate aspartate shuttle and glycerol phosphate shuttle, accomplish their transport (Fig. 14.14 ).
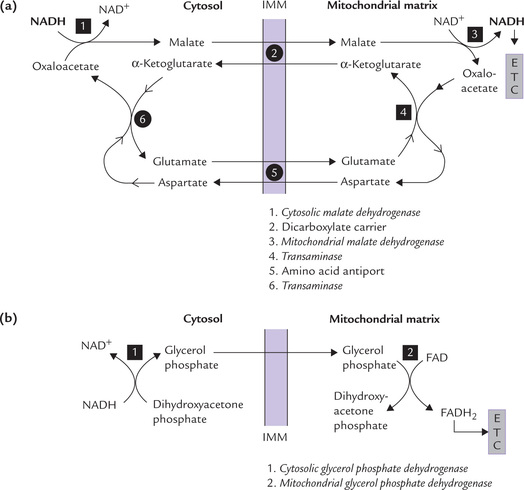
Fig. 14.14 Transfer of reducing equivalents from cytosol into mitochondrial matrix. (a) Malate aspartate shuttle, (b) Glycerol phosphate shuttle (IMM = mitochondrial matrix).
Malate aspartate shuttle: It operates in liver and heart muscles in the following steps:
1. The reducing equivalents are transferred from NADH to oxaloacetate to form malate. The reaction is catalyzed by the cytosolic enzyme malate dehydrogenase.
2. Malate is then transported across the IMM by the dicarboxylate carrier.
3. Within the mitochondrial matrix, the reducing equivalents are transferred from malate to NAD+ to form NADH. The enzyme that catalyzes this reaction is mitochondrial malate dehydrogenases.
Thus, by the concerted action of the dicarboxylate carrier and two enzymes (i.e. cytosolic malate dehydrogenase and mitochondrial malate dehydrogenase), NADH reaches the mitochondrial matrix (Fig. 14.14a). The mitochondrial NADH now contains the reducing equivalents that were originally present in the cytosolic NADH.
Rest of the shuttle systems is concerned with transport of the oxaloacetate back into the cytosol. It requires participation of amino acid antiport system which exchanges aspartate for glutamate across the IMM.
Glycerol phosphate shuttle: In skeletal muscle and brain, another type of shuttle, called glycerol phosphate shuttle, is present. It transfers the hydrogen first to dihydroxyacetone phosphate forming glycerol phosphate (in cytosol), and then to the FAD prosthetic group of the mitochondrial glycerol phosphate dehydrogenase (Fig. 14.14b). The latter is an integral protein of the inner mitochondrial membrane, which regenerates its FAD by direct transfer of electrons to the respiratory chain. Only two ATPs are therefore produced for each pair of reducing equivalents transferred.
IV Enzymes Participating in Biological Oxidation
The enzymes involved in biological oxidation catalyze oxidation-reduction reactions, hence they belong to the class I, i.e. oxidoreductases (Chapter 6). They are subclassified as: (a) dehydrogenases, (b) oxidases, and (c) hydroperoxidases.
A Dehydrogenases
They are most common of all oxidoreductases. They catalyze transfer of hydrogen between a substrate and a coenzyme, most commonly NAD+, FAD, NADP+ or FMN. These enzymes are named according to the substrate from which they remove the hydrogen (Table 14.1).
B Oxidases
They catalyze transfer of hydrogen removed from a substrate to oxygen. Mostly this results in formation of water. Cytochrome oxidase, the terminal component of ETC, is an example.
C Hydroperoxidases
They use hydrogen peroxide or an inorganic peroxide as a substrate. They are needed for degradation of hydrogen peroxide that may be produced in the reactions catalyzed by the dehydrogenases. Technically, catalase is also a peroxidase: it degrades hydrogen peroxide to molecular oxygen and water.
V Mitochondrial Myopathies
Mitochondrial DNA consists of a circular DNA molecule of 16,569 base pairs which encodes 13 polypeptides, 22 tRNAs, and the RNAs of mitochondrial ribosomes (12S and 16S). The mtDNA is maternally inherited since mitochondria from sperm does not enter the ovum during fertilization. The polypeptide chains encoded by mtDNA include seven subunits of NADH-Q reductase, one subunit of QH2-cytochrome c reductase, three subunits of cytochrome oxidase and two subunits of ATP synthase. All other mitochondrial proteins are encoded by nuclear genes and synthesized in cytoplasmic ribosomes.
Mitochondrial DNA is 10 times more susceptible to mutations than nuclear DNA. Some of these mutations result in absence of specific polypeptide chains. Faulty operation of the electron transport pathway results and the ATP production is decreased. These disorders are known as mitochondrial myopathies and they most commonly affect the tissues with high rate of oxidative phosphorylation. The mitochondrial myopathies are tissue specific; some affect the heart, others the skeletal muscles. Many are accompanied by lactic acidosis, because the inability to reduce NADH normally results in channeling of pyruvate toward lactic acid production (Chapter 9).
Leber’s hereditary optic neuropathy (LHON) is a mitochondrial myopathy, characterized by sudden onset of blindness in adults, associated with degeneration of the optic nerve.
Exercises
Essay type questions
1. What is redox potential and what is its significance? Describe various components of electron transport chain and discuss the oxidation of NADH and FADH2.
2. What do you understand by the term P : O ratio? Indicate diagrammatically the sites of ATP generation in the mitochondrial respiratory chain and their inhibitors.
3. What is oxidative phosphorylation? Discuss in detail the chemiosmotic hypothesis of ATP synthesis and explain how the process of oxidative phosphorylation is arrested by various inhibitors.
4. What are uncouplers of biological oxidation? State their mechanism of action.
 Respiratory chain or electron transport chain located in inner mitochondrial membrane consists of a series of electron carriers. Electrons from the reduced coenzymes, NADH and FADH2, pass through these before they reduce oxygen.
Respiratory chain or electron transport chain located in inner mitochondrial membrane consists of a series of electron carriers. Electrons from the reduced coenzymes, NADH and FADH2, pass through these before they reduce oxygen.