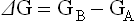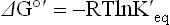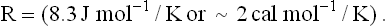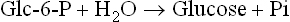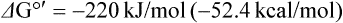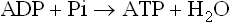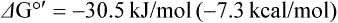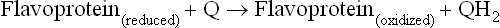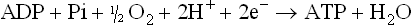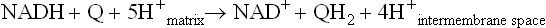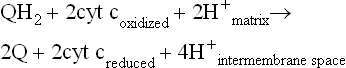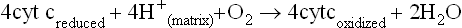Bioenergetics and Oxidative Metabolism
Introduction
ATP is the central metabolic currency
Oxidation of metabolic fuels is essential to life. In higher organisms, fuels such as carbohydrates and lipids are metabolized to carbon dioxide and water, generating a central metabolic currency, adenosine triphosphate (ATP). Most metabolic energy is produced by oxidation-reduction (redox) reactions in mitochondria. The regulation of energy metabolism is no small feat, because warm-blooded animals have such variable demands for energy from such processes as thermogenesis at low temperatures, stimulation of ATP synthesis during stress, and coupling of ATP synthesis with the rate of respiration during work and exercise. This chapter will provide an introduction to the concept of free energy, oxidative phosphorylation and the transduction of energy from fuels into useful work. The pathways and specific molecules through which electrons are transported to oxygen and the mechanism of generation of ATP will be described and related to the structure of the mitochondrion, the powerhouse of the cell and the major source of cellular ATP. Lastly, these biochemical processes will be applied to human health and disease.
Oxidation as a source of energy
Energy content of foods
Nutrition and disorders such as obesity, diabetes, and cancer all require an understanding of thermodynamics. Obesity, for example, is a disorder in which there is an imbalance between energy intake and expenditure. It is therefore important that the energy content of foods be known. The commonly accepted energy values for the four major food categories are shown in Table 9.1; alcohol is included because it is a significant dietary component for some people. These values are obtained by completely burning (oxidizing) samples of each food. Biologically, about 40% of food energy is conserved as ATP, and the remaining 60% is liberated as heat.
Table 9.1
Energy content of the major classes of food
| Metabolic fuel (kJ/g) | Energy content (kcal/g) | |
| Fats | 38 | 9 |
| Carbohydrates | 17 | 4 |
| Proteins | 17 | 4 |
| Alcohol | 29 | 7 |
Note that the thermodynamic term, kcal (energy required to increase the temperature of 1 kg (1 L) of water by 1°C) is equivalent to the common nutritional Calorie (capital C), i.e. 1 Cal = 1 kcal, 1 kcal = 4.2 kJ.
The basal metabolic rate (BMR)
The basal metabolic rate is a measure of the total daily energy expenditure by the body at rest
Virtually all of the reactions in the body are exothermic, and the sum of all reactions at rest is called the basal metabolic rate (BMR), which can be measured by two basic methods: direct calorimetry, where the total heat liberated by an animal is measured over time, and indirect calorimetry, where the BMR is calculated from the quantity of oxygen consumed, which is directly related to the BMR. Adult men (70 kg) have a BMR of about 7500 kJ (1800 kcal) and women about 5400 kJ (1300 kcal) per day; the BMR may vary by a factor of two between individuals, depending on age, sex, body mass and composition. Heat production by mitochondria accounts for the largest portion of the BMR. The BMR is measured under controlled conditions: after an 8-hour sleep, in the reclining position, in the postabsorptive state, typically after a 12-hour fast.
Another measure often used is the RMR, or resting metabolic rate, which is virtually the same as the BMR but measured under less restrictive conditions. The RMR is a measure of minimum energy expenditure at rest; it is typically about 70% of total daily energy expenditure. Exercise scientists frequently use the term MET, metabolic equivalent task, as a measure of energy expenditure at rest. Slow to vigorous walking is a 2–4 MET/h activity; vigorous running on a treadmill may consume more than 15 MET/h.
Stages of fuel oxidation
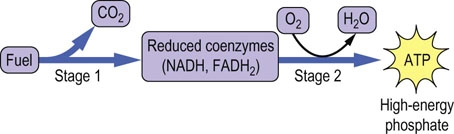
The oxidation of fuels can be divided into two general stages: production of reduced nucleotide coenzymes during the oxidation of fuels, and use of the free energy from oxidation of the reduced coenzymes to produce ATP (Fig. 9.1).
Free energy
The direction of a reaction depends on the difference between the free energy of reactants and products
The Gibbs' free energy (ΔG) of a reaction is the maximum amount of energy that can be obtained from a reaction at constant temperature and pressure. The units of free energy are kcal/mol (kJ/mol). It is not possible to measure the absolute free energy content of a substance directly, but when reactant A reacts to form product B, the free energy change in this reaction, ΔG, can be determined.
where GA and GB are the free energy of A (reactant) and B (product), respectively. All reactions in biological systems are considered to be reversible reactions, so that the free energy of the reverse reaction, B → A, is numerically equivalent, but opposite in sign to that of the forward reaction.
If there is a greater concentration of B than of A at equilibrium, i.e. Keq > 1, the conversion A → B is favorable – that is, the reaction tends to move forward from a standard state in which A and B are present at equal concentrations. In this case, the reaction is said to be a spontaneous or exergonic reaction, and the free energy of this reaction is defined as negative: that is, ΔG < 0, indicating that energy is liberated by the reaction. Conversely, if the concentration of A is greater than that of B at equilibrium, the forward reaction is termed unfavorable, nonspontaneous or endergonic, and the reaction has a positive free energy: that is, when starting concentrations are equal, B tends to form A, rather than A to form B. In this case, energy input would be required to push the reaction A → B forward from its equilibrium position to the standard state in which A and B are present at equal concentrations. The total free energy available from a reaction depends on both its tendency to proceed forward from the standard state (ΔG) and the amount (moles) of reactant converted to product.
The free energy of metabolic reactions is related to their equilibrium constants by the Gibbs' equation
Thermodynamic measurements are based on standard-state conditions where reactant and product are present at 1 molar concentrations, the pressure of all gases is 1 atmosphere and the temperature is 25°C (298K). Most commonly, the concentrations of reactants and products are then measured after equilibrium is attained. Standard free energies are represented by the symbol ΔG° and biological standard free energy change by ΔG°′, with the accent symbol designating pH 7.0. The free energy available from a reaction may be calculated from its equilibrium constant by the Gibbs' equation:
where T is absolute temperature (Kelvin), lnK′eq is the natural logarithm of the equilibrium constant for the reaction at pH 7, and R is the ideal gas constant:
Several common metabolic intermediates that you will encounter in your studies are listed in Table 9.2, along with the equilibrium constants and free energies for their hydrolysis reactions. Those intermediates with free energy changes equal to or greater than that of ATP, the central energy transducer of the cell, are considered to be high-energy compounds, and generally have either anhydride or thioester bonds. The lower-energy compounds listed are all phosphate esters and, in comparison, do not yield as much free energy on hydrolysis. The hydrolysis reaction of glucose-6-phosphate (Glc-6-P) is written as:
Table 9.2
Thermodynamics of hydrolysis reactions
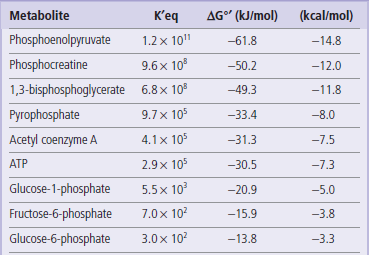
Equilibrium constants and free energy of hydrolysis of various metabolic intermediates at pH 7 (ΔG°′).
This reaction has a negative free energy and occurs spontaneously. The reverse reaction, synthesis of Glc-6-P from glucose and phosphate, would require input of energy.
Conservation of energy by coupling with adenosine triphosphate
ATP is a product of catabolic reactions and a driver of biosynthetic reactions
Living systems must transfer energy from one molecule to another without losing all of it as heat. Some of the energy must be conserved in a chemical form in order to drive nonspontaneous biosynthetic reactions. In fact, nearly half of the energy obtained from the oxidation of metabolic fuels is channeled into the synthesis of ATP, a universal energy transducer in living systems. ATP is often referred to as the common currency of metabolic energy, because it is used to drive so many energy-requiring reactions. ATP consists of the purine base adenine, the five-carbon sugar ribose, and α, β, and γ phosphate groups (Fig. 9.2). The two phosphoanhydride linkages are said to be high-energy bonds, because their hydrolysis yields a large negative change in free energy. When ATP is used for metabolic work, these high-energy linkages are broken and ATP is converted to ADP or to AMP.
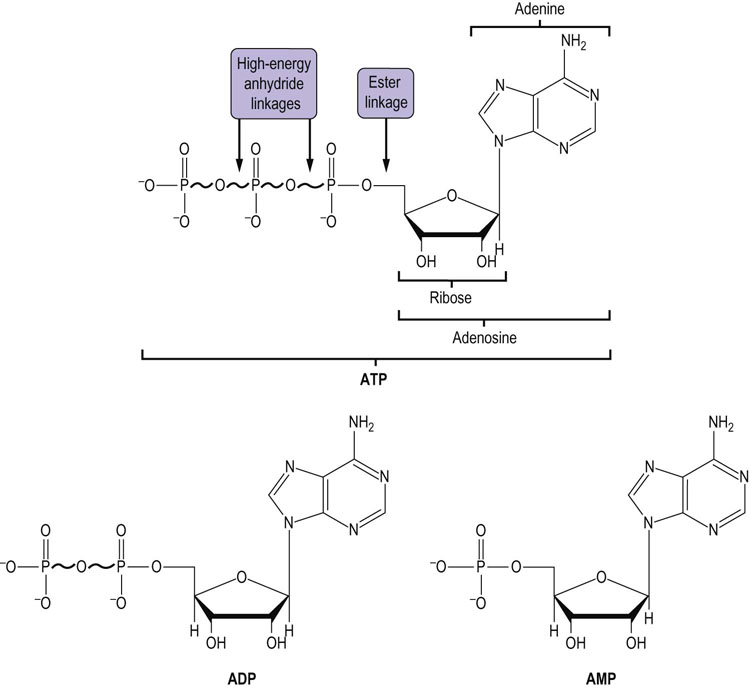
Fig. 9.2 Structures of adenine nucleotides.
ATP is shown, together with its hydrolysis products, adenosine diphosphate (ADP) and adenosine monophosphate (AMP). ATP has two high-energy phosphoanhydride bonds; ADP has one; and AMP has only a low-energy phosphoester bond.
The free energy of a high-energy bond, such as the phosphate anhydride bonds in ATP, can be used to drive or push forward reactions that would otherwise be unfavorable. In fact, nearly all biosynthetic pathways are thermodynamically unfavorable, but are made favorable by coupling various reactions with hydrolysis of high-energy compounds. For example, the first step in the metabolism of glucose is the synthesis of Glc-6-P (Fig. 3.4). As shown in Table 9.2, this is not a favorable reaction: the hydrolysis of Glc-6-P (ΔG°′ = −13.8 kJ/mol or −3.3 kcal/mol) is the favored reaction. However, as shown below, the synthesis of Glc-6-P (reaction I) can be energetically coupled to the hydrolysis of ATP (reaction II), yielding a ‘net reaction’ III that is favorable for synthesis of Glc-6-P:
This is possible because of the high free energy or ‘group transfer potential’ of ATP. The physical transfer of the phosphate from ATP to glucose occurs in the active site of a kinase enzyme, such as glucokinase. This motif, in which ATP is used to drive biosynthetic reactions, transport processes or muscle activity, occurs commonly in metabolic pathways.
Mitochondrial synthesis of adenosine triphosphate from reduced coenzymes
Oxidative phosphorylation is the mechanism by which energy derived from fuel oxidation is conserved in the form of ATP
Metabolism of carbohydrates begins in the cytoplasm through the glycolytic pathway (see Chapter 12), whereas energy production from fatty acids occurs exclusively in the mitochondrion. Mitochondria are subcellular organelles, about the size of bacteria. They are essential for aerobic metabolism in eukaryotes. Their main function is to oxidize metabolic fuels and conserve free energy by synthesizing ATP.
Mitochondria are bounded by a dual membrane system (Fig. 9.3). The outer membrane (OMM) contains enzyme and transport proteins and via the pore-forming protein porin (P), it is permeable to virtually all ions, small molecules (S) and proteins less than 10,000 Da. Large proteins must be transported via the TOM (translocase in the outer mitochondrial membrane) and TIM (translocase in the inner mitochondrial membrane) complexes. This is especially vital to the cell, because almost all mitochondrial proteins are nuclear encoded and must be transported into the mitochondrion. The mitochondrial genome, mtDNA, encodes 13 vital subunits of the proton pumps and ATP synthase. The inner membrane (IMM) is pleated with structures known as cristae, and is impermeable to most ions and small molecules, such as nucleotides (including ATP), coenzymes, phosphate, and protons. Transporter proteins are required to selectively facilitate translocation of specific molecules across the inner membrane. The inner membrane also contains components of oxidative phosphorylation – the process by which the oxidation of reduced nucleotide coenzymes is coupled to the synthesis of ATP.
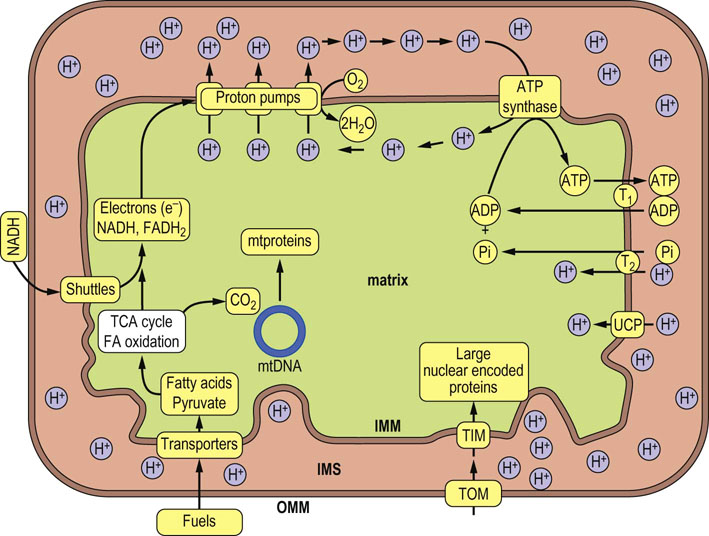
Fig. 9.3 Mitochondrial structure and pathways of energy transduction: the mechanism of oxidative phosphorylation.
Major fuels, such as pyruvate from carbohydrates and fatty acids (FA) from triglycerides, are transported into the matrix where they are oxidized to generate CO2 and the reduced nucleotide coenzymes NADH and FADH2. Oxidation of these nucleotides via the electron transport system reduces oxygen to water and pumps protons by three proton pumps out of the matrix and into the intermembrane space (IMS), creating a pH gradient, which is the major contributor to the membrane potential. It should be noted that protons in the intermembrane space freely diffuse through the outer membrane via the protein porin, a proton channel, so the intermembrane space is roughly equivalent to the cytosol. Although the membrane potential is mostly composed of the proton gradient it actually consists of several electrochemical gradients and is expressed as a voltage. Controlled influx of protons through ATP synthase powers the synthesis of ATP by ATP synthase (F-ATPase, Table 8.3). Mitochondrial ATP is then exchanged for cytoplasmic ADP through the ADP-ATP translocase (T1). Phosphate (Pi), which is also required for ATP synthesis, is transported by the phosphate translocase (T2). The inner membrane also contains uncoupling proteins (UCP) that may be used to allow the controlled leakage of protons back into the matrix. OMM, outer mitochondrial membrane; IMM, inner mitochondrial membrane; mtproteins, mitochondrial proteins; mtDNA, mitochondrial DNA; TOM and TIM, protein translocase complexes in outer and inner mitochondrial membrane; TCA, tricarboxylic acid cycle.
Transduction of energy from reduced coenzymes to high-energy phosphate
NAD+, FAD, and FMN are the major redox coenzymes
The major redox coenzymes involved in transduction of energy from fuels to ATP are nicotinamide adenine dinucleotide (NAD+), flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN) (Fig. 9.4). During energy metabolism, electrons are transferred from carbohydrates and fats to these coenzymes, reducing them to NADH, FADH2 and FMNH2. In each case, two electrons are transferred, but the number of protons transferred differs. NAD+ accepts a hydride ion (H–) that consists of one proton and two electrons; the remaining proton is released into solution. FAD and FMN accept two electrons and two protons.
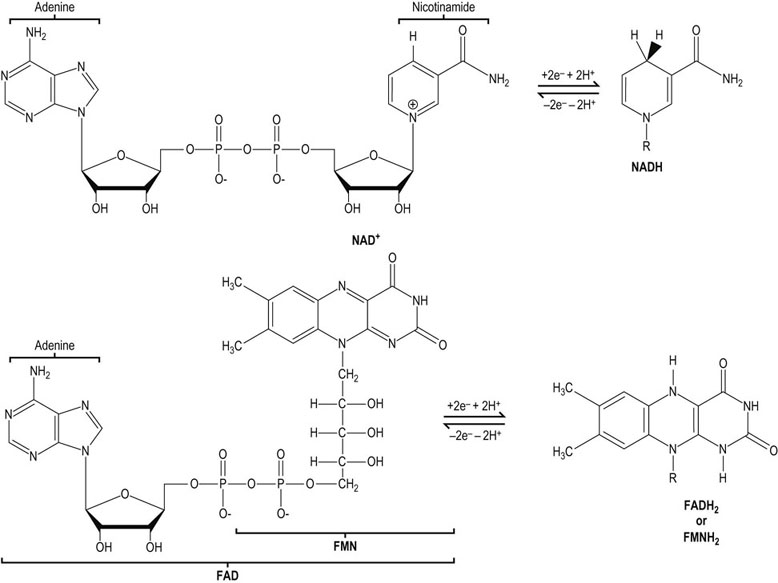
Fig. 9.4 The structure of redox coenzymes.
NAD+ and its reduced form, NADH (nicotinamide adenine dinucleotide), consists of adenine, two ribose units, two phosphates, and nicotinamide. FAD and its reduced form, FADH2 (flavin adenine dinucleotide), consists of riboflavin, two phosphates, ribose and adenine. FMN and FMNH2 consist of riboflavin phosphate. The nicotinamide and riboflavin components of these coenzymes are reversibly oxidized and reduced during electron transfer (redox) reactions. NADH and FADH2 are often called reduced nucleotides or reduced coenzymes.
The oxidation of reduced nucleotides by the electron transport system produces a large amount of free energy. When the oxidation of 1 mole of NADH is coupled to the reduction of 0.5 mole of oxygen to form water, the energy produced is theoretically sufficient to synthesize 7 moles of ATP:
Dividing 220 kJ/mol of ΔG°′ available from oxidation of NADH by ΔG°′ 30.5 required for synthesis of ATP yields, theoretically, 7 mol ATP/mol NADH. As discussed below, the actual yield is closer to 2.5 mol ATP/mol NADH oxidized.
The free energy of oxidation of NADH and FADH2 is used by the electron transport system to pump protons into the intermembrane space. The energy produced when these protons reenter the mitochondrial matrix is used to synthesize ATP. This process is known as oxidative phosphorylation (see Fig. 9.3)
The mitochondrial electron transport system
The mitochondrial electron transport chain transfers electrons in a defined multi-step sequence from reduced nucleotides to oxygen
The entire electron transport system, also known as the electron transport chain or respiratory chain, is located in the inner mitochondrial membrane (Fig. 9.5). It consists of several large protein complexes and two small, independent components – ubiquinone and cytochrome c. The protein components are very complex; complex I, for example, which accepts electrons from NADH, contains at least 46 subunits. Each step in the electron transport chain involves a redox reaction where electrons are transferred from components with more negative reduction potentials to components with more positive reduction potentials. Electrons are conducted through this system in a defined sequence from reduced nucleotide coenzymes to oxygen, and the free energy changes drive the transport of protons from the matrix into the intermembrane space via the three proton pumps. After each step, the electrons are at a lower energy state.
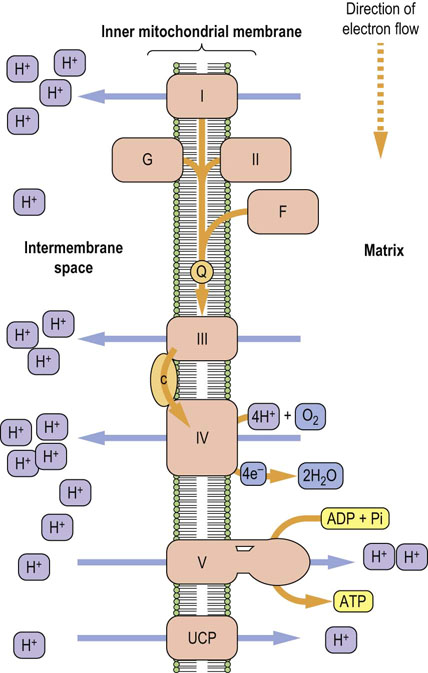
Fig. 9.5 A section of the mitochondrial inner membrane with the electron transport system and ATP synthase.
I, complex I; II, complex II (succinate dehydrogenase); III, complex III; IV, complex IV; V, complex V or ATP synthase; G, glycerol-3-phosphate dehydrogenase; F, fatty acyl CoA dehydrogenase; Q, ubiquinone; c, cytochrome c; UCP, uncoupling protein.
Electrons are funneled into the electron transport chain by several flavoproteins
There are four flavoproteins in the electron transport chain: Complex I contains FMN and the other three contain FAD. These flavoproteins all reduce the small, lipophilic molecule ubiquinone (Q or coenzyme Q10), at the beginning of the common electron transport pathway, consisting of Q, complex III, cytochrome c, and complex IV.
Protons are pumped from the matrix into the intermembrane space by complexes I, III, and IV. Oxygen (O2) is the final electron acceptor at the end of the chain, and it is reduced to two water molecules by the transfer of four electrons from complex IV and four protons from the mitochondrial matrix compartment.
The efficiency of oxidative phosphorylation can be measured by dividing the amount of phosphate incorporated into ADP by the amount of atomic oxygen reduced. One atom of oxygen is reduced by two electrons (one electron pair).
For each pair of electrons transported through complexes I, III or IV, a sufficient number of protons is pumped by each complex for the synthesis of approximately one mole of ATP/complex. If electron transport begins with an electron pair from NADH, approximately 2.5 moles of ATP are synthesized, whereas an electron pair from any of the other three FADH2-containing flavoproteins yields about 1.5 moles of ATP, because the proton-pumping capability of complex I is bypassed.
Flavoproteins contain FAD or FMN prosthetic groups
Complex I, also called NADH–Q reductase or NADH dehydrogenase, is a flavoprotein containing FMN. It oxidizes mitochondrial NADH, and transfers electrons through FMN and iron–sulfur (FeS) complexes to ubiquinone, providing enough energy to pump four protons from the matrix in the reaction:
Three other flavoproteins transfer electrons from oxidizable substrates via FADH2 to ubiquinone (Q) (see Fig. 9.5):
 Succinate – Q reductase (complex II or succinate dehydrogenase of the TCA cycle) (see Chapter 14) oxidizes succinate to fumarate and reduces FAD to FADH2.
Succinate – Q reductase (complex II or succinate dehydrogenase of the TCA cycle) (see Chapter 14) oxidizes succinate to fumarate and reduces FAD to FADH2.
 Glycerol-3-phosphate – Q reductase, a part of the glycerol-3-P shuttle (see below), oxidizes cytoplasmic glycerol-3-P to dihydroxyacetone phosphate (DHAP) and reduces FAD to FADH2.
Glycerol-3-phosphate – Q reductase, a part of the glycerol-3-P shuttle (see below), oxidizes cytoplasmic glycerol-3-P to dihydroxyacetone phosphate (DHAP) and reduces FAD to FADH2.
 Fatty acyl CoA dehydrogenase catalyzes the first step in the mitochondrial oxidation of fatty acids and also produces FADH2.
Fatty acyl CoA dehydrogenase catalyzes the first step in the mitochondrial oxidation of fatty acids and also produces FADH2.
Both FMN and FAD contain the water-soluble vitamin riboflavin. A dietary deficiency of riboflavin can severely impair the function of these and other flavoproteins.
Transfer of electrons from NADH into mitochondria
Electron shuttles
Electron shuttles are required for mitochondrial oxidation of NADH produced in the cytoplasmic compartment
NADH is produced in the cytosol during carbohydrate metabolism. NADH cannot cross the inner mitochondrial membrane, and therefore it cannot be oxidized by the electron transport system. Two redox shuttles permit the oxidation of cytosolic NADH without its physical transfer into the mitochondrion. A characteristic feature of these shuttles is that they are powered by cytoplasmic and mitochondrial isoforms of the same enzyme, which catalyze opposing reactions on opposite sides of the membrane. The glycerol-3-P shuttle is the simpler of the two (Fig. 9.7, top). It transfers the electrons of NADH from the cytoplasm to the mitochondrion by reducing FAD to FADH2. Cytoplasmic glycerol-3-P dehydrogenase catalyzes reduction of dihydroxyacetone-P (DHAP) with NADH to glycerol-3-P, regenerating NAD+. The cytoplasmic glycerol-3-phosphate is oxidized back to DHAP by another glycerol-3-phosphate dehydrogenase isoform facing the outer surface of the inner mitochondrial membrane; this enzyme is a flavoprotein in which FAD is reduced to FADH2. The electrons are then transferred to the common pathway via ubiquinone. Because the electrons are transferred to FAD, the yield of ATP from cytoplasmic NADH by this pathway is approximately 1.5 moles, rather than the 2.5 moles available from mitochondrial NADH via the NADH-Q reductase complex (complex I).
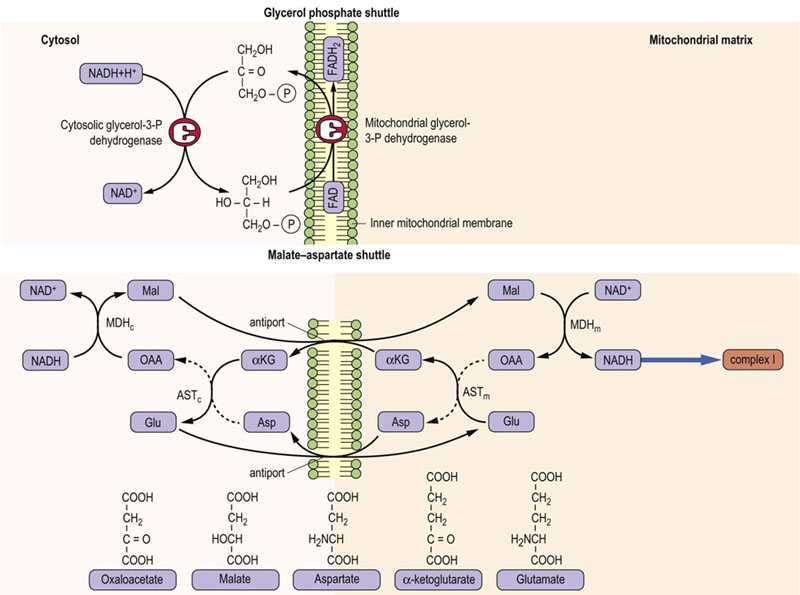
Fig. 9.7 Redox shuttles in the inner mitochondrial membrane.
(Top) The glycerol phosphate shuttle. (Bottom) The malate–aspartate shuttle MDH, malate dehydrogenase; AST, aspartate aminotransferase. Subscripts c and m refer to cytosolic and mitochondrial isozymes.
Many cells, e.g. in skeletal muscle, use the glycerol-3-P shuttle, but heart and liver rely on the malate–aspartate shuttle (Fig. 9.7, bottom), which yields 2.5 moles of ATP per mole of NADH. This shuttle is more complicated, because the substrate, malate, can cross the inner mitochondrial membrane, but the membrane is impermeable to the product, oxaloacetate – there is no oxaloacetate transporter. The exchange is therefore accomplished by interconversion between α-keto- and α-amino acids, involving cytoplasmic and mitochondrial glutamate and α-ketoglutarate, and isozymes of glutamate-oxaloacetate transaminase (aspartate aminotransferase).
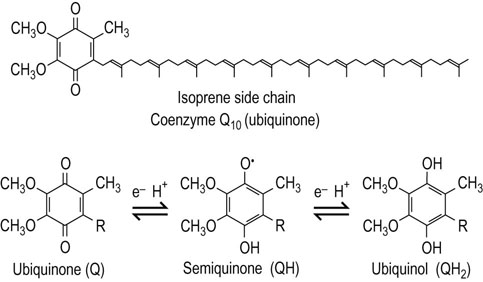
Ubiquinone (coenzyme Q10)
Ubiquinone tranfers electrons from flavoproteins to complex III
Ubiquinone is so named because it is ubiquitous in virtually all living systems. It is a small, lipid-soluble compound found in the inner membrane of animal and plant mitochondria and in the plasma membrane of bacteria. The primary form of mammalian ubiquinone contains a side chain of 10 isoprene units and is often called CoQ10. It diffuses within the inner membrane, accepts electrons from the four major mitochondrial flavoproteins, and transfers them to complex III (QH2–cytochrome c reductase). Ubiquinone can carry either one or two electrons (Fig. 9.8) and is also thought to be a major source of superoxide radicals in the cell (see Chapter 37).
Complex III – cytochrome c reductase
Complex III accepts electrons from ubiquinone and pumps four hydrogen ions across the inner mitochondrial membrane
This enzyme complex, also known as ubiquinone–cytochrome c reductase or QH2–cytochrome c reductase, oxidizes ubiquinone and reduces cytochrome c. Reduced ubiquinone funnels electrons that it gathers from mitochondrial flavoproteins and transfers them to complex III. Electrons from ubiquinone are transferred through two species of cytochrome b, to an FeS center, to cytochrome c1, and finally to cytochrome c. Transport of two electrons to cytochrome c yields sufficient free energy change and protons pumped to synthesize about one mole of ATP. The overall reaction is:
Four protons are pumped during this reaction, two from fully reduced ubiquinone and two from the matrix.
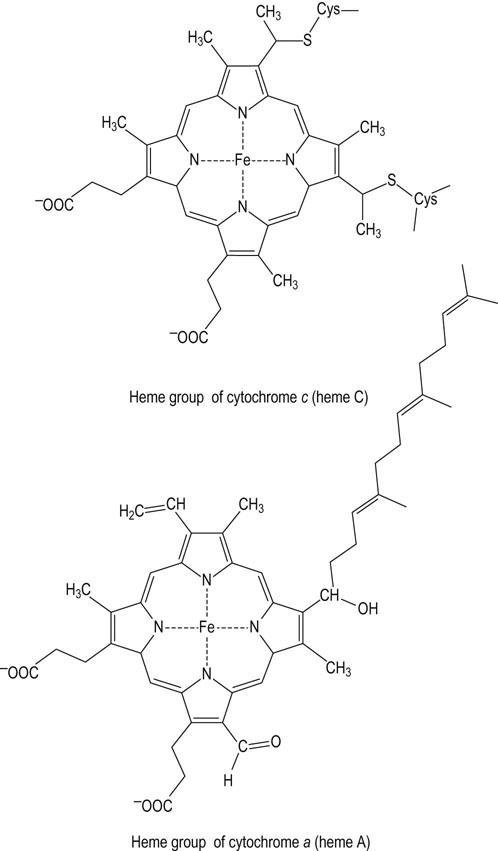
Cytochrome c
Cytochrome c is a peripheral membrane proteins, shuttling electrons from complex III to complex IV
Cytochrome c, a small heme protein that is loosely bound to the outer surface of the inner membrane, shuttles electrons from complex III to complex IV. Each cytochrome c carries only one electron, so the reduction of O2 to 2H2O by complex IV requires four reduced cytochrome c molecules. The binding of cytochrome c to complexes III and IV is largely electrostatic, involving a number of lysine residues on the protein surface. Reduction of ferricytochrome c (Fe3+) to ferrocytochrome c (Fe2+) by cytochrome c1 leads to a change in the three-dimensional structure, charge distribution and dipole moment of the protein, promoting transfer of electrons to cytochrome a in complex IV (see Fig. 9.5). In response to oxidative stress and cell injury (Chapter 37), cytochrome c may be released from the inner mitochondrial membrane and leak into the cytosol, inducing apoptosis (cell death).
Complex IV
Complex IV, at the end of the electron transport chain, transfers electrons to oxygen, producing water
Complex IV, known as cytochrome c oxidase or cytochrome oxidase, exists as a dimer in the IMM. It oxidizes the mobile cytochrome c, and conducts electrons through cytochromes a and a3, finally reducing oxygen to water in a four-electron transfer reaction (Fig. 9.10). Copper is a common component of this and other oxidase enzymes. Small molecule poisons, such as azide, cyanide, and carbon monoxide, bind to the heme group of cytochrome a3 in cytochrome c oxidase and inhibit complex IV. In common with complexes I and III, the cytochrome oxidase complex pumps protons out of mitochondria, providing for the synthesis of about one mole of ATP per pair of electrons transferred to oxygen. The actual number of protons pumped is four. In addition, another four are required in the reduction of O2 to water. The overall reaction catalyzed by complex IV is:

Fig. 9.10 Complex IV.
Complex IV utilizes four electrons from cytochrome c and eight protons from the matrix. Four protons and electrons reduce oxygen to water. Four additional protons are pumped out of the matrix. Complex IV is regulated allosterically by ATP, by reversible phosphorylation/dephosphorylation, and by thyroid hormone (T2 or diiodothyronine). a, cytochrome a; a3, cytochrome a3.
Synthesis of adenosine triphosphate – the chemiosmotic hypothesis
According to the chemiosmotic hypothesis, mitochondria produce ATP using the free energy from the proton gradient generated during oxidation of NADH and FADH2. This energy is described as a proton motive force, an electrochemical gradient created by the proton concentration gradient and a proton charge differential (outside positive) across the inner mitochondrial membrane. To operate, it requires an inner membrane system that is impermeable to protons, except through ATP synthase or other complexes in a regulated fashion. When protons are pumped out of the matrix, the intermembrane space becomes more acidic and more positively charged than the matrix.
The ATP synthase complex (complex V) is an example of rotary catalysis
Lining the inner matrix face of the inner membrane of each mitochondrion are thousands of copies of the ATP synthase complex, also called complex V or F0F1-ATP synthase (F = coupling factor; see Inhibitors of ATP synthase, below). ATP synthase is also called an ATPase, because it can hydrolyze ATP. ATP synthase consists of two major complexes (Fig. 9.11). The inner membrane component, termed F0, is the proton-driven motor with the stoichiometry of a, b2 and c10–14. The c-subunits form the c-ring, which rotates in a clockwise direction in response to the flow of protons through the complex. Since the γ- and ε-subunits are bound to the c-ring, they rotate with it, inducing large conformational changes in the three-αβ dimers of the F1 complex. The two β-proteins immobilize the second complex (F1-ATP synthase).
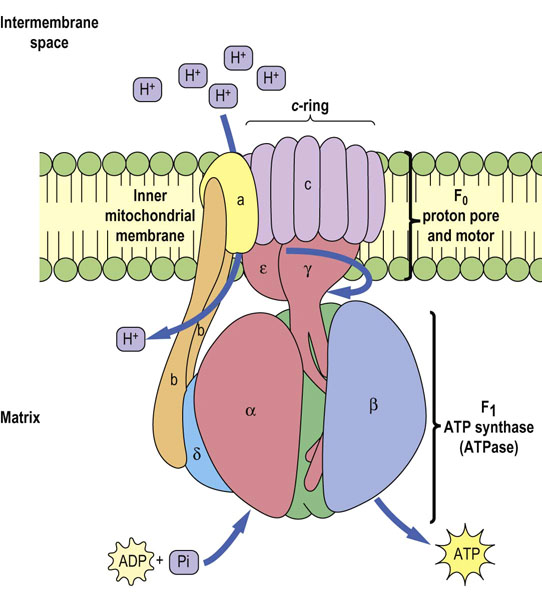
Fig. 9.11 ATP synthase complex.
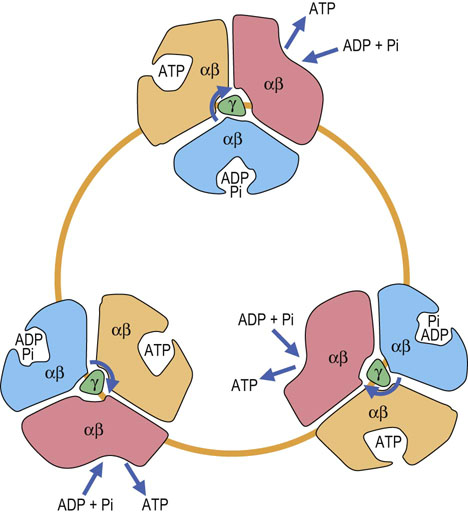
The ATP synthase complex consists of a motor (F0) and generator (F1). The proton pore involves the c-ring and the a-protein. The rotary component is the coiled-coil γ-subunit, which is bound to the ε-subunit and to the c-ring. The stationary component is the hexameric α3β3 unit, which is held in place by the δ, b and a-proteins.
F1 has a stoichiometry of α3, β3, γ, δ, ε. The major part of F1 consists of three αβ dimers arranged like slices of an orange, with the catalytic activity residing on the β-subunits. Each 120° rotation of the γ-subunit induces conformational changes in the αβ-dimeric subunits such that the nucleotide-binding sites alternate between three states: the first binds ADP and Pi, the second synthesizes ATP, and the third releases ATP, so each complete turn produces 3ATP. This is known as the binding-change mechanism (Fig. 9.12). Surprisingly, the proton-motive free energy used by ATP synthase is not for ATP synthesis itself, but for its release; when the proton gradient is too low to support ATP release, ATP remains stuck to ATP synthase and further ATP production ceases. ADP and Pi are bound to the complex as soon as ATP leaves. The αβ-dimers are asymmetrical, because each is in a different conformation at any given moment. This complex is a proton-driven motor, and it is an example of rotary catalysis. About three protons are required for the synthesis of each ATP. This complex acts independently of the electron transport chain; addition of a weak acid, such as acetic acid, to a suspension of isolated mitochondria is sufficient to induce the biosynthesis of ATP in vitro.
P : O ratios
The P : O ratio is a measure of the number of high-energy phosphates (i.e. amount of ATP) synthesized per atom of oxygen (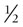 O2) consumed, or per mole of water produced. The P : O ratio can be calculated from the moles of ADP used to synthesize ATP and the atoms of oxygen taken up by mitochondria. For example, if 2 mmol of ADP is converted to ATP and 0.5 mmol of oxygen (1.0 milliatom of oxygen) is taken up, the P : O ratio is 2.0. As discussed earlier, the theoretical yield of ATP per mole of NADH is about 7 moles; however, by actual measurement with isolated mitochondria, the P : O ratio for oxidation of metabolites that yield NADH is about 2.5 and the ratio for those that yield FADH2 is about 1.5. The remainder of the energy available from the oxidation of NADH and FADH2 is released in the form of heat.
O2) consumed, or per mole of water produced. The P : O ratio can be calculated from the moles of ADP used to synthesize ATP and the atoms of oxygen taken up by mitochondria. For example, if 2 mmol of ADP is converted to ATP and 0.5 mmol of oxygen (1.0 milliatom of oxygen) is taken up, the P : O ratio is 2.0. As discussed earlier, the theoretical yield of ATP per mole of NADH is about 7 moles; however, by actual measurement with isolated mitochondria, the P : O ratio for oxidation of metabolites that yield NADH is about 2.5 and the ratio for those that yield FADH2 is about 1.5. The remainder of the energy available from the oxidation of NADH and FADH2 is released in the form of heat.
‘Respiratory control’ is the dependence of oxygen uptake by mitochondria on the availability of ADP
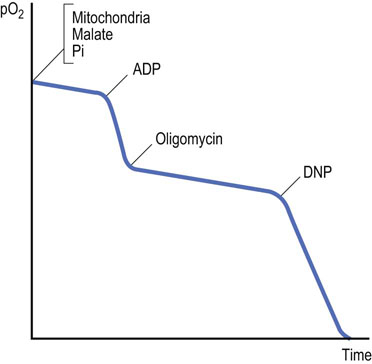
Normally, oxidation and phosphorylation are tightly coupled: substrates are oxidized, electrons are transported, and oxygen is consumed only when synthesis of ATP is required (coupled respiration). Thus, resting mitochondria consume oxygen at a slow rate, which can be greatly stimulated by addition of ADP (Fig. 9.13). ADP is taken up by the mitochondria and stimulates ATP synthase, which lowers the proton gradient. Respiration increases, because the proton pumps are stimulated to reestablish the proton gradient. When the ADP is depleted, ATP synthesis terminates and respiration returns to the original rate. Oxygen uptake declines to the original rate when the concentration of ADP is depleted and ATP synthesis terminates.
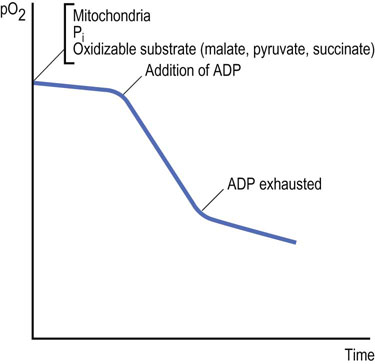
Fig. 9.13 Effect of ADP on the uptake of oxygen by isolated mitochondria.
This may be studied in an isolated (sealed) system with an oxygen electrode and a recording device. The graph shows a typical recording of oxygen consumption (pO2, partial pressure of oxygen) by normal mitochondria on introduction of ADP.
Mitochondria can become partially uncoupled if the inner membrane loses its structural integrity. They are said to be ‘leaky’, because protons can diffuse through the inner membrane without involving ATP synthase. This occurs if isolated mitochondria are treated with mild detergents that disrupt the inner membrane, or if they have been stored for a period of time. Such mitochondria are said to be ‘uncoupled’; oxidation proceeds without production of ATP and uncoupled mitochondria lose respiratory control because protons pumped by the electron chain bypass the ATPase and leak unproductively back into the matrix. The P : O ratio declines under these conditions.
The mechanism of respiratory control depends on the requirement for ADP and Pi binding to the ATP synthase complex: in the absence of ADP and Pi, protons cannot enter the mitochondrion through this complex and oxygen consumption markedly decreases, because the proton pumps cannot pump protons against a high proton back-pressure. This happens because the free energy of the electron transport reactions is sufficient to generate a pH gradient of only two units across the membrane. If the pH gradient cannot be discharged for production of ATP, the two pH unit gradient is established and the pumps grind to a halt and stall. The electron transport chain becomes reduced and substrate oxidation and oxygen consumption decrease. A little physical activity, with consumption of ATP and generation of ADP and Pi, opens up the ATPase channels, discharging the proton gradient and activating the electron transport chain, and fuel and oxygen consumption. At a whole-body level, we breathe faster during exercise to provide the additional oxygen needed for increased oxidative phosphorylation.
Uncouplers
Uncouplers and uncoupling proteins are thermogenic
Uncouplers of oxidative phosphorylation dissipate the proton gradient by transporting protons back into mitochondria, bypassing the ATP synthase. Uncouplers stimulate respiration, because the system makes a futile attempt to restore the proton gradient by oxidizing more fuel and pumping more protons out of mitochondria. Uncouplers are typically hydrophobic compounds and either weak acids or bases, with pKa near pH 7. The classic uncoupler, 2,4-dinitrophenol (DNP) (Fig. 9.14), is protonated in solution on the outer, more acidic side of the inner mitochondrial membrane. Because of its hydrophobicity, it may then freely diffuse through the inner mitochondrial membrane. When it reaches the matrix side, it encounters a more basic pH and the proton is released, effectively discharging the pH gradient. Other uncouplers include preservatives and antimicrobial agents, such as pentachlorophenol and p-cresol.

Fig. 9.14 Proton transport by uncouplers.
Uncouplers transport protons into the mitochondrion, dissipating the proton gradient. DNP is an example of an exogenous uncoupler. The uncoupling proteins (UCP) are endogenous uncouplers in the IMM and are regulated by hormones. The gradient consisting of protons and other factors constitute the mitochondrial membrane potential (MMP), which is expressed in millivolts (mV). DNP, 2,4-dinitrophenol; IMM, inner mitochondrial membrane.
Uncoupling proteins (UCP)
According to the chemiosmotic hypothesis, the inner mitochondrial membrane is topologically closed. However, it has long been known that protons are transported into the matrix from the intermembrane space by routes other than the ATP synthase complex and inner membrane transporters. Much of the BMR is now thought to be mainly due to inner membrane components called uncoupling proteins (UCP). The first discovered was uncoupling protein-1 (UCP1), formerly known as thermogenin, which is found exclusively in brown adipose tissue. Brown adipose tissue is abundant in the newborn and in some adult mammals, and it is brown because of its high content of mitochondria. In humans, brown adipose tissue is abundant in infants, but it gradually diminishes and is barely detectable in adults. UCP1 provides body heat during cold stress in the young and in some adult animals. It accomplishes this by uncoupling the proton gradient, thereby generating heat (thermogenesis) instead of ATP. Uncoupling proteins are expressed at high levels in hibernating animals, permitting them to maintain body temperature without movement or exercise.
Four additional uncoupling proteins are expressed by the human genome: UCP2, UCP3, UCP4 and UCP5. While UCP1 is exclusive to brown adipose tissue, UCP2 is expressed ubiquitously, UCP3 is mainly expressed in skeletal muscle, and UCP4 and UCP5 are expressed in the brain. Except for UCP1, the physiologic functions of these proteins are not well understood, but could be of profound significance in our understanding of such health issues as diabetes, obesity, cancer, thyroid disease and aging. As uncouplers, they have been linked to a number of fundamental functions. For example, there is strong evidence that obesity induces the synthesis of UCP2 in β-cells of the pancreas. This may play a role in the β-cell dysfunction found in type 2 diabetes, because it lowers the intracellular concentration of ATP, which is required for secretion of insulin. The thyroid hormone (T3) has been shown to stimulate thermogenesis in rats by promoting the synthesis of UCP3 in skeletal muscle. The common fever that is induced by infectious organisms is probably also due to uncoupling by the UCP, but the mechanism is unknown.
Inhibitors of oxidative metabolism
Electron transport system inhibitors
Inhibitors of electron transport selectively inhibit complexes I, III or IV, interrupting the flow of electrons through the respiratory chain. This stops proton pumping, ATP synthesis, and oxygen uptake. Several inhibitors are readily available poisons that could be encountered in the practice of medicine. It is noteworthy that genetic defects in respiratory chain components often mimic the effects of these inhibitors.
Rotenone inhibits complex I (NADH–Q reductase)
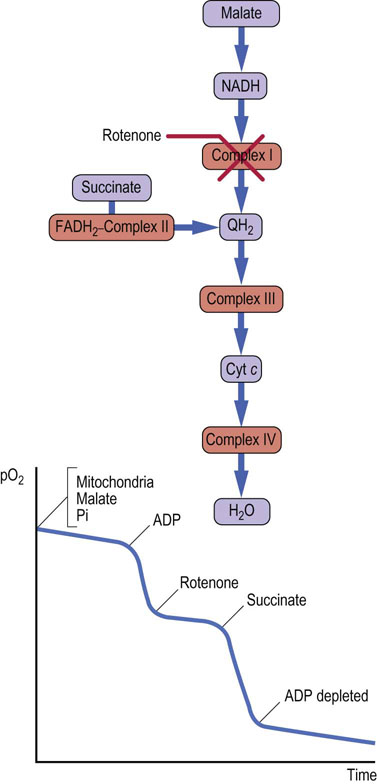
Rotenone, a common insecticide, and some barbiturates (e.g. amytal) inhibit complex I. Because malate and lactate are oxidized by NAD+, their oxidation will be decreased by rotenone. However, substrates yielding FADH2 can still be oxidized, because complex I is bypassed and electrons are donated to ubiquinone from FADH2. Addition of ADP to a suspension of mitochondria supplemented with malate and phosphate (Fig. 9.15) markedly stimulates oxygen uptake as ATP synthesis occurs. Oxygen uptake is markedly inhibited by rotenone, but when succinate is added, ATP synthesis and oxygen consumption resume until the supply of ADP is exhausted. Rotenone inhibition of complex I causes reduction of all components prior to the point of inhibition, because they cannot be oxidized, whereas those after the point of inhibition become fully oxidized. This is known as a crossover point, and it can be determined spectrophotometrically, because light absorption by respiratory chain components changes according to redox state. Such analyses were used to define the sequence of components in the respiratory chain.
Antimycin A inhibits complex III (QH2–cytochrome c reductase)
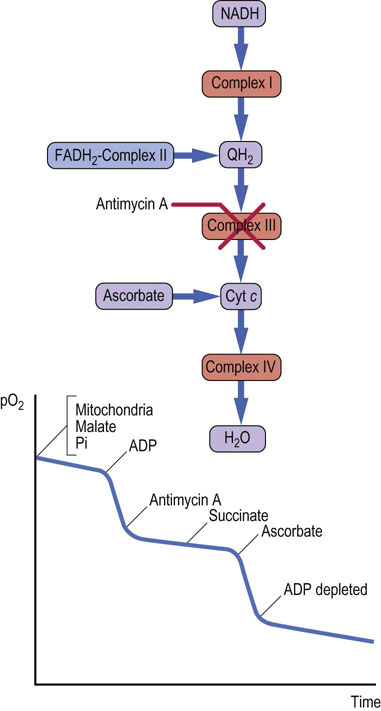
The inhibition of complex III by antimycin A prevents transfer of electrons from either complex I or FADH2-containing flavoproteins to cytochrome c. In this case, components preceding complex III become fully reduced, and those after it become oxidized. The oxygen uptake curve (Fig. 9.16) shows that the stimulation of respiration by ADP is inhibited by antimycin A, but that the addition of succinate does not relieve the inhibition. Ascorbic acid can reduce cytochrome c, and addition of ascorbic acid restores respiration, illustrating that complex IV is unaffected by antimycin A.
Cyanide and carbon monoxide inhibit complex IV
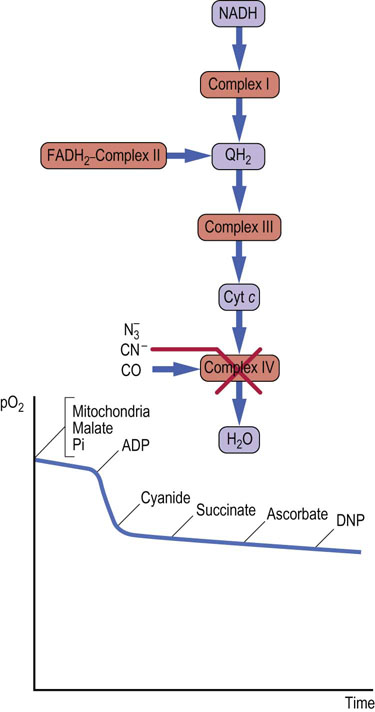
Azide 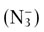 , cyanide (CN)−; and carbon monoxide (CO) inhibit complex IV (cytochrome c oxidase) (Fig. 9.17). Because complex IV is the terminal electron transfer complex, its inhibition cannot be bypassed. All components preceding complex IV become reduced, oxygen cannot be reduced, none of the complexes can pump protons, and ATP is not synthesized. Uncouplers such as DNP have no effect, because there is no proton gradient. Cyanide and carbon monoxide also bind to hemoglobin, blocking oxygen binding and transport (see Chapter 5). In these poisonings, both the ability to transport oxygen and to synthesize ATP are impaired. The administration of oxygen is used for the treatment of such poisonings. It is of interest that sodium azide is the nitrogen source for the inflation of air bags; it may pose an environmental problem if it is accidentally released, i.e. nonexplosively.
, cyanide (CN)−; and carbon monoxide (CO) inhibit complex IV (cytochrome c oxidase) (Fig. 9.17). Because complex IV is the terminal electron transfer complex, its inhibition cannot be bypassed. All components preceding complex IV become reduced, oxygen cannot be reduced, none of the complexes can pump protons, and ATP is not synthesized. Uncouplers such as DNP have no effect, because there is no proton gradient. Cyanide and carbon monoxide also bind to hemoglobin, blocking oxygen binding and transport (see Chapter 5). In these poisonings, both the ability to transport oxygen and to synthesize ATP are impaired. The administration of oxygen is used for the treatment of such poisonings. It is of interest that sodium azide is the nitrogen source for the inflation of air bags; it may pose an environmental problem if it is accidentally released, i.e. nonexplosively.
Oligomycin inhibits ATP synthase
Oligomycin inhibits respiration but, in contrast to electron transport inhibitors, it is not a direct inhibitor of the electron transport system. Instead, it inhibits the proton channel of ATP synthase. It causes an accumulation of protons outside the mitochondrion, because the proton pumping system is still intact but the proton channel is blocked. The addition of the uncoupler DNP after oxygen uptake has been inhibited by oligomycin illustrates this point: DNP dissipates the proton gradient and stimulates oxygen uptake as the electron transport system attempts to reestablish the proton gradient (Fig. 9.18).
Inhibitors of the ADP–ATP translocase
Most ATP is synthesized in the mitochondrion, but used in the cytosol for biosynthetic reactions. Newly synthesized mitochondrial ATP and spent cytosolic ADP are exchanged by a mitochondrial ADP–ATP translocase, representing about 10% of the protein in the inner mitochondrial membrane (see Fig. 9.3). This translocase can be inhibited by unusual plant and mold toxins, such as bongkrekic acid and atractyloside. Their effects are similar to those of oligomycin in vitro – a proton gradient builds up and electron transport stops, but, as with oligomycin, respiration can be reactivated by uncouplers.
Regulation of oxidative phosphorylation
Respiratory control and feedback regulation
ADP is the key feedback regulator of oxidative phosphorylation
The oldest and simplest known mechanism of respiratory control is accomplished by the supply of ADP. This is based on the fact that when added to isolated mitochondria, ADP stimulates respiration and ATP synthesis. When ADP is completely converted to ATP, respiration returns to the initial rate. Oxidative phosphorylation is also tightly coupled to other fundamental pathways such as glycolysis, fatty acid oxidation and the tricarboxylic acid cycle (see Chapters 12, 14 and 15) through feedback regulatory mechanisms. The ratios of NADH/NAD and ATP/ADP feedback on key enzymes, such as PFK-1, pyruvate dehydrogenase and isocitrate dehydrogenase. If, for example, oxidative phosphorylation is switched off because of high ATP, both NADH and ATP will negatively feedback on other extramitochondrial pathways, limiting the flux of fuel to mitochondria. Since oxidative phosphorylation depends on the supply of FADH2, NADH, ADP, and Pi as well as the ATP/ADP ratio, magnitude of the membrane potential, uncoupling and hormonal factors, its modes of regulation are clearly complex.
Regulation by covalent modification and allosteric effectors (ATP–ADP)
The main target for regulating oxidative phosphorylation appears to be complex IV. It is phosphorylated in response to hormone action (see Chapter 13) by cyclic-3′5′-adenosine monophosphate (cAMP)-dependent protein kinase (PKA) and dephosphorylated by a Ca2+-stimulated protein phosphatase. Phosphorylation enables allosteric regulation by ATP (ATP/ADP ratio). A high ATP/ADP ratio inhibits and a low ratio stimulates oxidative phosphorylation. It is thought that the complex is normally phosphorylated and inhibited by ATP. With high Ca2+ levels, e.g. in muscle during exercise (see Chapter 20), the enzyme is dephosphorylated, the inhibition by ATP is abolished, and its activity greatly stimulated, increasing ATP.
Based on the observation that in type 2 diabetes, ATP production is decreased when the β-subunit of ATP synthase is phosphorylated, it has been proposed that this complex is also regulated by phosphorylation/dephosphorylation. Note that the secretion of insulin in β-cells of the pancreas is ATP dependent, because ATP binds to the ATP-sensitive potassium channel (see Insulin Secretion, Chapter 21).
Regulation by thyroid hormones
Thyroid hormones act at two levels in mitochondria. In rats, T3 stimulates the synthesis of UCP2 and UCP3, which can uncouple the proton gradient, but this has not been documented in humans. Additionally, T2 binds to complex IV on the matrix side, inducing slip in cytochrome c oxidase. The term ‘slip’ means that complex IV pumps fewer protons per electron transported through the complex, resulting in thermogenesis. The action of T3 could explain, in part, the long-term and T2 the short-term thermogenic effects of thyroid hormones (see also Chapter 39).
Mitochondrial permeability transition pore (MPTP)
Located in the inner mitochondrial membrane, the MPTP is a nonselective pore that is a critical factor in cell death. It is normally closed, but will open when cells are reperfused after a period of ischemia (ischemic reperfusion injury, IRI; Chapter 37), and small molecules will leave the mitochrondrial matrix. Opening of the MPTP is now considered a key feature of IRI in which cellular damage is much greater than that produced by ischemia alone. Cascades of reactions occuring in response to IRI lead to apoptosis and necrosis, and cell death.
Ischemia, such as that found in heart attacks, is usually caused by a clot that blocks an artery. Clot busters such as streptokinase can be administered to dissolve clots and reperfuse ischemic cells. But, if the ischemic state has been prolonged before administration of a clot buster, death may result from reperfusion injury and opening of the MPTP. This occurs all too often in heart attack patients. Several drugs, such as cyclosporin A, inhibit the MPTP from opening and may protect cells from necrosis or apoptosis after the administration of a clot buster. In fact, a large clinical trial of cyclosporin A is currently being conducted with heart attack patients in conjunction with the administration of a clot buster.
Summary
 The electron transport system consists of electron carriers that are located in the inner mitochondrial membrane, each of which is isolable as a complex or as a single molecule.
The electron transport system consists of electron carriers that are located in the inner mitochondrial membrane, each of which is isolable as a complex or as a single molecule.
 Electrons from four major flavoproteins feed electrons to ubiquinone, the first member of the common pathway. Energy derived from the conductance of electrons through the electron transport system is used by three of the complexes to pump protons into the intermembrane space, creating an electrochemical gradient or proton motive force.
Electrons from four major flavoproteins feed electrons to ubiquinone, the first member of the common pathway. Energy derived from the conductance of electrons through the electron transport system is used by three of the complexes to pump protons into the intermembrane space, creating an electrochemical gradient or proton motive force.
 The proton gradient is used to power ATP synthase for synthesis of ATP by rotary catalysis as well as transport of intermediates across the inner membrane.
The proton gradient is used to power ATP synthase for synthesis of ATP by rotary catalysis as well as transport of intermediates across the inner membrane.
 Numerous toxins can severely impair the electron transport system, the ATP synthase and the translocase that exchanges ATP and ADP across the inner mitochondrial membrane.
Numerous toxins can severely impair the electron transport system, the ATP synthase and the translocase that exchanges ATP and ADP across the inner mitochondrial membrane.
 The rate of ATP production by the electron transport system is regulated by modulation of the proton gradient, by allosteric modification and phosphorylation–dephosphorylation and by thyroid hormones.
The rate of ATP production by the electron transport system is regulated by modulation of the proton gradient, by allosteric modification and phosphorylation–dephosphorylation and by thyroid hormones.
 At least five uncoupling proteins (UCP) with specific tissue distributions are found in the inner mitochondrial membrane, and they all regulate the membrane potential, energy expenditure and thermogenesis.
At least five uncoupling proteins (UCP) with specific tissue distributions are found in the inner mitochondrial membrane, and they all regulate the membrane potential, energy expenditure and thermogenesis.
 Chronic diseases or conditions such as diabetes, cancer, obesity, and aging all have metabolic links to dysregulation of oxidative phosphorylation through effects on the electron transport system and ATP synthase.
Chronic diseases or conditions such as diabetes, cancer, obesity, and aging all have metabolic links to dysregulation of oxidative phosphorylation through effects on the electron transport system and ATP synthase.
 The integrity of mitochondria and cells may be disrupted by ischemic reperfusion injury and opening of the mitochondrial permeability transition pore, leading to death and tissue damage.
The integrity of mitochondria and cells may be disrupted by ischemic reperfusion injury and opening of the mitochondrial permeability transition pore, leading to death and tissue damage.
Cao, XH, Zhao, SS, Liu, DY, et al. ROS-Ca(2+) is associated with mitochondria permeability transition pore involved in surfactin-induced MCF-7 cells apoptosis. Chem Biol Interact. 2011; 190:16–27.
Picard, M, Taivassalo, T, Gouspillou, G, et al. Mitochondria: isolation, structure and function. J Physiol. 2011; 589:4413–4421.
Pinadda, V, Halestrap, AP. The roles of phosphate and the phosphate carrier in the mitochondrial permeability transition pore. Mitochondrion. 2012; 12:120–125.
Ramsden, DB, Ho, PW, Ho, JW, et al. Human neuronal uncoupling proteins 4 and 5 (UCP4 and UCP5): structural properties, regulation, and physiological role in protection against oxidative stress and mitochondrial dysfunction. Brain Behav. 2012; 2:468–478.
Ruiz-Meana, M, Fernandez-Sanz, C, Garcia-Dorado, D. The SR-mitochondria interaction: a new player in cardiac pathophysiology. Cardiovasc Res. 2010; 88:30–39.
Shanbhag, R, Shi, G, Rujiviphat, J, et al. The emerging role of proteolysis in mitochondrial quality control and the etiology of Parkinson's disease. Parkinsons Dis. 2012; 2012:382175.
Vonck, J, Schäfer, E. Supramolecular organization of protein complexes in the mitochondrial inner membrane. Biochim Biophys Acta. 2009; 1793:117–124.
Wang, WH, Chiang, IT, Ding, K, et al. Curcumin-induced apoptosis in human hepatocellular carcinoma j5 cells: critical role of Ca(2+)-dependent pathway. Evid Based Complement Alternat Med. 2012; 2012:512907.
www.cnr.berkeley.edu/~hongwang/Project/ATP_synthase/
www.youtube.com/watch?v=PjdPTY1wHdQ
ATP synthas. http://vcell.ndsu.nodak.edu/animations/atpgradient/index.htm
Virtual Cell Animation Cente. http://vcell.ndsu.nodak.edu/animations/home.htm
Bioenergetic. www.bmb.leeds.ac.uk/illingworth/oxphos/
The Children's Mitochondrial Disease Networ. www.emdn-mitonet.co.uk/
United Mitochondrial disease Foundatio. www.umdf.org/mitodisease/