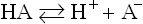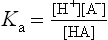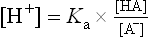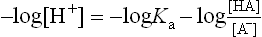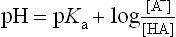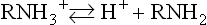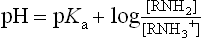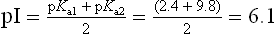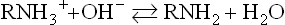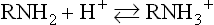Amino Acids and Proteins
Introduction
Proteins are major structural and functional polymers in living systems
Proteins have a broad range of activities, including catalysis of metabolic reactions and transport of vitamins, minerals, oxygen, and fuels. Some proteins make up the structure of tissues, while others function in nerve transmission, muscle contraction and cell motility, and still others in blood clotting and immunologic defenses, and as hormones and regulatory molecules. Proteins are synthesized as a sequence of amino acids linked together in a linear polyamide (polypeptide) structure, but they assume complex three-dimensional shapes in performing their function. There are about 300 amino acids present in various animal, plant and microbial systems, but only 20 amino acids are coded by DNA to appear in proteins. Many proteins also contain modified amino acids and accessory components, termed prosthetic groups. A range of chemical techniques is used to isolate and characterize proteins by a variety of criteria, including mass, charge and three-dimensional structure. Proteomics is an emerging field which studies the full range of expression of proteins in a cell or organism, and changes in protein expression in response to growth, hormones, stress, and aging.
Amino acids
Amino acids are the building blocks of proteins
Stereochemistry: configuration at the α-carbon, D- and L-isomers
Each amino acid has a central carbon, called the α-carbon, to which four different groups are attached (Fig. 2.1):
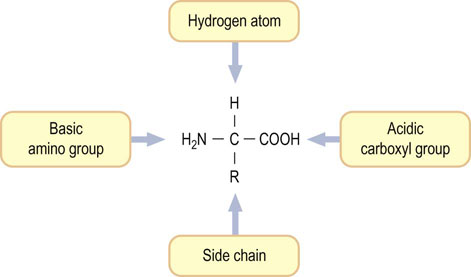
Fig. 2.1 Structure of an amino acid.
Except for glycine, four different groups are attached to the α-carbon of an amino acid. Table 2.1 lists the structures of the R groups.
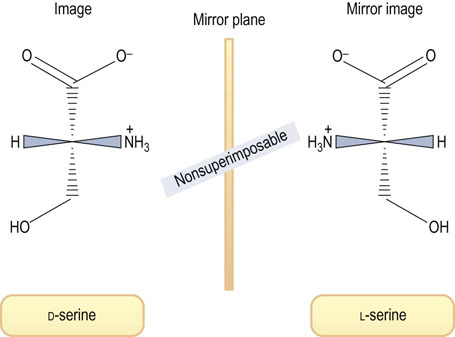
One of the 20 amino acids, proline, is not an α-amino acid but an α-imino acid (see below). Except for glycine, all amino acids contain at least one asymmetric carbon atom (the α-carbon atom), giving two isomers that are optically active, i.e. they can rotate plane-polarized light. These isomers, referred to as stereoisomers or enantiomers, are said to be chiral, a word derived from the Greek word for hand. Such isomers are nonsuperimposable mirror images and are analogous to left and right hands, as shown in Figure 2.2. The two amino acid configurations are called D (for dextro or right) and L (for levo or left). All amino acids in proteins are of the L-configuration, because proteins are biosynthesized by enzymes that insert only L-amino acids into the peptide chains.
Classification of amino acids based on chemical structure of their side chains
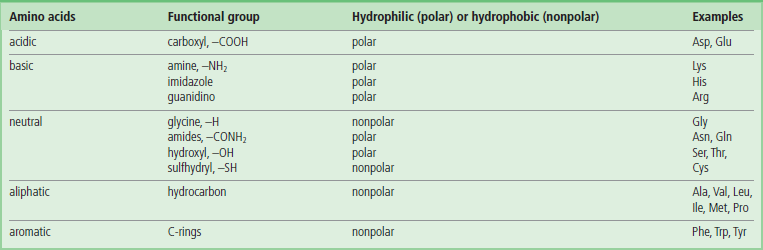
The properties of each amino acid are dependent on its side chain ( R), which determines; the side chains are the functional groups that the structure and function of proteins, as well as the electrical charge of the molecule. Knowledge of the properties of these side chains is important for understanding methods of analysis, purification, and identification of proteins. Amino acids with charged, polar or hydrophilic side chains are usually exposed on the surface of proteins. The nonpolar hydrophobic residues are usually buried in the hydrophobic interior or core of a protein and are out of contact with water. The 20 amino acids in proteins encoded by DNA are listed in Table 2.1 and are classified according to their side chain functional groups.
R), which determines; the side chains are the functional groups that the structure and function of proteins, as well as the electrical charge of the molecule. Knowledge of the properties of these side chains is important for understanding methods of analysis, purification, and identification of proteins. Amino acids with charged, polar or hydrophilic side chains are usually exposed on the surface of proteins. The nonpolar hydrophobic residues are usually buried in the hydrophobic interior or core of a protein and are out of contact with water. The 20 amino acids in proteins encoded by DNA are listed in Table 2.1 and are classified according to their side chain functional groups.
Table 2.1
The 20 Amino Acids found in proteins.*
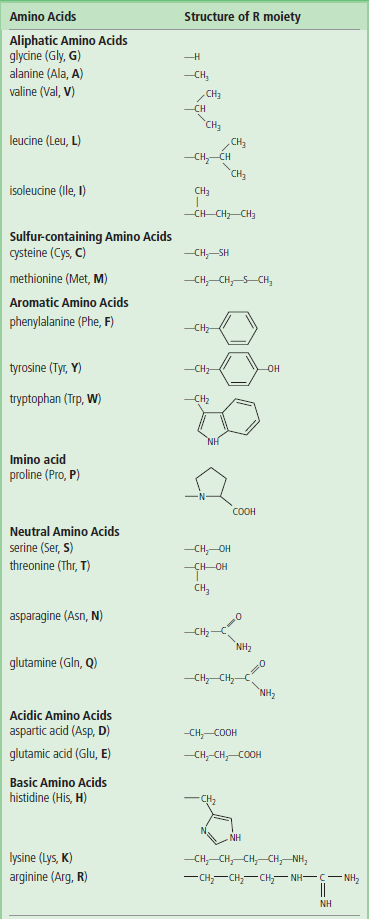
*The three-letter and single-letter abbreviations in common use are given in parentheses.
Aliphatic amino acids
Alanine, valine, leucine, and isoleucine, referred to as aliphatic amino acids, have saturated hydrocarbons as side chains. Glycine, which has only a hydrogen side chain, is also included in this group. Alanine has a relatively simple structure, a side chain methyl group, while leucine and isoleucine have sec- and iso-butyl groups. All of these amino acids are hydrophobic in nature.
Aromatic amino acids
Phenylalanine, tyrosine, and tryptophan have aromatic side chains
The nonpolar aliphatic and aromatic amino acids are normally buried in the protein core and are involved in hydrophobic interactions with one another. Tyrosine has a weakly acidic hydroxyl group and may be located on the surface of proteins. Reversible phosphorylation of the hydroxyl group of tyrosine in some enzymes is important in the regulation of metabolic pathways. The aromatic amino acids are responsible for the ultraviolet absorption of most proteins, which have absorption maxima ~280 nm. Tryptophan has a greater absorption in this region than the other two aromatic amino acids. The molar absorption coefficient of a protein is useful in determining the concentration of a protein in solution, based on spectrophotometry. Typical absorption spectra of aromatic amino acids and a protein are shown in Figure 2.3.
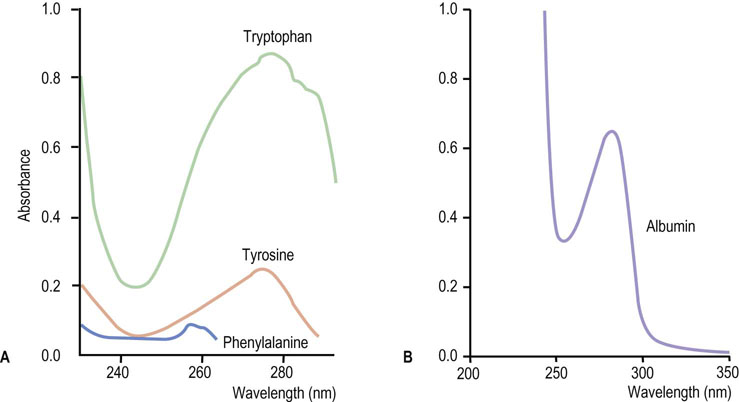
Fig. 2.3 Ultraviolet absorption spectra of the aromatic amino acids and bovine serum albumin.
(A) Aromatic amino acids such as tryptophan, tyrosine, and phenylalanine have absorbance maxima at ∼280 nm. Each purified protein has a distinct molecular absorption coefficient at around 280 nm, depending on its content of aromatic amino acids. (B) A bovine serum albumin solution (1 mg dissolved in 1 mL of water) has an absorbance of 0.67 at 280 nm using a 1 cm cuvette. The absorption coefficient of proteins is often expressed as E1% (10 mg/mL solution). For albumin, E1%280 nm = 6.7. Although proteins vary in their Trp, Tyr, and Phe content, measurements of absorbance at 280 nm are useful for estimating protein concentration in solutions.
Neutral polar amino acids
Neutral polar amino acids contain hydroxyl or amide side chain groups. Serine and threonine contain hydroxyl groups. These amino acids are sometimes found at the active sites of catalytic proteins, enzymes (Chapter 6). Reversible phosphorylation of peripheral serine and threonine residues of enzymes is also involved in regulation of energy metabolism and fuel storage in the body (Chapter 13). Asparagine and glutamine have amide-bearing side chains. These are polar but uncharged under physiologic conditions. Serine, threonine and asparagine are the primary sites of linkage of sugars to proteins, forming glycoproteins (Chapter 26).
Acidic amino acids
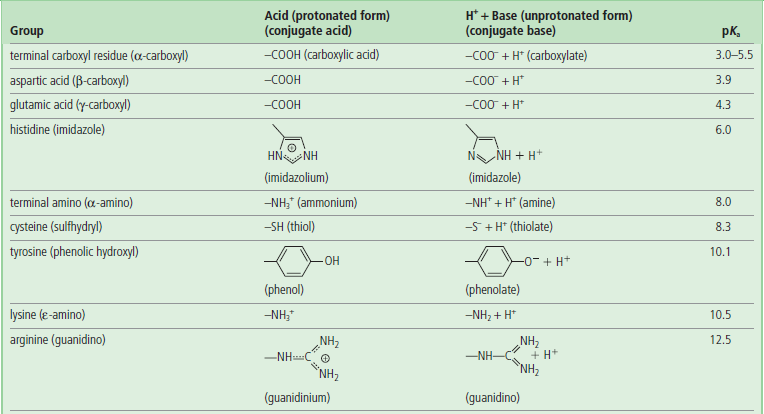
Aspartic and glutamic acids contain carboxylic acids on their side chains and are ionized at pH 7.0 and, as a result, carry negative charges on their β- and γ-carboxyl groups, respectively. In the ionized state, these amino acids are referred to as aspartate and glutamate, respectively.
Basic amino acids
The side chains of lysine and arginine are fully protonated at neutral pH and, therefore, positively charged. Lysine contains a primary amino group (NH2) attached to the terminal ε-carbon of the side chain. The ε-amino group of lysine has a pKa ≈ 11. Arginine is the most basic amino acid (pKa ≈ 13) and its guanidine group exists as a protonated guanidinium ion at pH 7.0.
Histidine (pKa ≈ 6) has an imidazole ring as the side chain and functions as a general acid–base catalyst in many enzymes. The protonated form of imidazole is called an imidazolium ion.
Sulfur-containing amino acids
Cysteine and its oxidized form, cystine, are sulfur-containing amino acids characterized by low polarity. Cysteine plays an important role in stabilization of protein structure, since it can participate in formation of a disulfide bond with other cysteine residues to form cystine residues, crosslinking protein chains and stabilizing protein structure. Two regions of a single polypeptide chain, remote from each other in the sequence, may be covalently linked through a disulfide bond (intrachain disulfide bond). Disulfide bonds are also formed between two polypeptide chains (interchain disulfide bond), forming covalent protein dimers. These bonds can be reduced by enzymes or by reducing agents such as 2-mercaptoethanol or dithiothreitol, to form cysteine residues. Methionine is the third sulfur-containing amino acid and contains a nonpolar methyl thioether group in its side chain.
Classification of amino acids based on the polarity of the amino acid side chains
Table 2.2 depicts the functional groups of amino acids and their polarity (hydrophilicity). Polar side chains can be involved in hydrogen bonding to water and to other polar groups and are usually located on the surface of the protein. Hydrophobic side chains contribute to protein folding by hydrophobic interactions and are located primarily in the core of the protein or on surfaces involved in interactions with other proteins.
Ionization state of an amino acid
Amino acids are amphoteric molecules – they have both basic and acidic groups
Monoamino and monocarboxylic acids are ionized in different ways in solution, depending on the solution's pH. At pH 7, the ‘zwitterion’ +H3N CH2
CH2 COO− is the dominant species of glycine in solution, and the overall molecule is therefore electrically neutral. On titration to acidic pH, the α-amino group is protonated and positively charged, yielding the cation +H3N
COO− is the dominant species of glycine in solution, and the overall molecule is therefore electrically neutral. On titration to acidic pH, the α-amino group is protonated and positively charged, yielding the cation +H3N CH2
CH2 COOH, while titration with alkali yields the anionic H2N
COOH, while titration with alkali yields the anionic H2N CH2
CH2 COO− species.
COO− species.
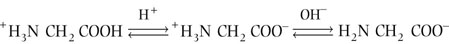
pKa values for the α-amino and α-carboxyl groups and side chains of acidic and basic amino acids are shown in Table 2.3. The overall charge on a protein depends on the contribution from basic (positive charge) and acidic (negative charge) amino acids, but the actual charge on the protein varies with the pH of the solution. To understand how the side chains affect the charge on proteins, it is worth recalling the Henderson–Hasselbalch equation.
Henderson–Hasselbalch equation and pKa
The H-H equation describes the titration of an amino acid and can be used to predict the net charge and isoelectric point of a protein
The general dissociation of a weak acid, such as a carboxylic acid, is given by the equation:
where HA is the protonated form (conjugate acid or associated form) and A− is the unprotonated form (conjugate base, or dissociated form).
The dissociation constant (Ka) of a weak acid is defined as the equilibrium constant for the dissociation reaction (1) of the acid:
The hydrogen ion concentration [H+] of a solution of a weak acid can then be calculated as follows. Equation (2) can be rearranged to give:
Equation (3) can be expressed in terms of a negative logarithm:
Since pH is the negative logarithm of [H+], i.e. −log[H+] and pKa equals the negative logarithm of the dissociation constant for a weak acid, i.e. −logKa, the Henderson–Hasselbalch equation (5) can be developed and used for analysis of acid–base equilibrium systems:
For a weak base, such as an amine, the dissociation reaction can be written as:
and the Henderson–Hasselbalch equation becomes:
From equations (5) and (7), it is apparent that the extent of protonation of acidic and basic functional groups, and therefore the net charge will vary with the pKa of the functional group and the pH of the solution. For alanine, which has two functional groups with pKa = 2.4 and 9.8, respectively (Fig. 2.4), the net charge varies with pH, from +1 to −1. At a point intermediate between pKa1 and pKa2, alanine has a net zero charge. This pH is called its isoelectric point, pI (Fig. 2.4).
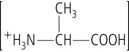
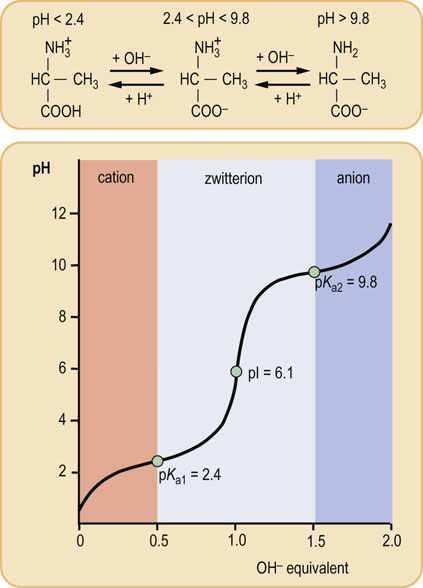
Fig. 2.4 Titration of amino acid.
The curve shows the number of equivalents of NaOH consumed by alanine while titrating the solution from pH 0 to pH 12. Alanine contains two ionizable groups: an α-carboxyl group and an α-amino group. As NaOH is added, these two groups are titrated. The pKa of the α-COOH group is 2.4, whereas that of the α-NH3+ group is 9.8. At very low pH, the predominant ion species of alanine is the fully protonated, cationic form:
At the mid-point in the first stage of the titration (pH 2.4), equimolar concentrations of proton donor and proton acceptor species are present, providing good buffering power.
At the mid-point in the overall titration, pH 6.1, the zwitterion is the predominant form of the amino acid in solution. The amino acid has a net zero charge at this pH – the negative charge of the carboxylate ion being neutralized by the positive charge of the ammonium group.
The second stage of the titration corresponds to the removal of a proton from the
 NH3+ group of alanine. The pH at the mid-point of this stage is 9.8, equal to the pKa for the
NH3+ group of alanine. The pH at the mid-point of this stage is 9.8, equal to the pKa for the  NH3+ group. The titration is complete at a pH of about 12, at which point the predominant form of alanine is the unprotonated, anionic form:
NH3+ group. The titration is complete at a pH of about 12, at which point the predominant form of alanine is the unprotonated, anionic form:The pH at which a molecule has no net charge is known as its isoelectric point, pI. For alanine, it is calculated as:
Buffers
Amino acids and proteins are excellent buffers under physiological conditions
Buffers are solutions that minimize a change in [H+], i.e. pH, on addition of acid or base. A buffer solution, containing a weak acid or weak base and a counter-ion, has maximal buffering capacity at its pKa, i.e. when the acidic and basic forms are present at equal concentrations. The acidic, protonated form reacts with added base, and the basic unprotonated form neutralizes added acid, as shown below for an amino compound:
An alanine solution (Fig. 2.4) has maximal buffering capacity at pH 2.4 and 9.8, i.e. at the pKa of the carboxyl and amino groups, respectively. When dissolved in water, alanine exists as a dipolar ion, or zwitterion, in which the carboxyl group is unprotonated ( COO−) and the amino group is protonated (
COO−) and the amino group is protonated ( NH3+). The pH of the solution is 6.1, the pI, half-way between the pKa of the amino and carboxyl groups. The titration curve of alanine by NaOH (Fig. 2.4) illustrates that alanine has minimal buffering capacity at its pI, and maximal buffering capacity at a pH equal to the pKa1 or pKa2.
NH3+). The pH of the solution is 6.1, the pI, half-way between the pKa of the amino and carboxyl groups. The titration curve of alanine by NaOH (Fig. 2.4) illustrates that alanine has minimal buffering capacity at its pI, and maximal buffering capacity at a pH equal to the pKa1 or pKa2.
Peptides and proteins
Primary structure of proteins
The primary structure of a protein is the linear sequence of its amino acids
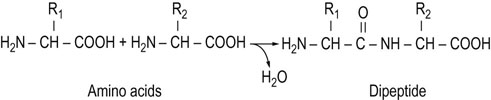
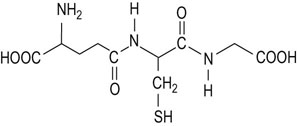
In proteins, the carboxyl group of one amino acid is linked to the amino group of the next amino acid, forming an amide (peptide) bond; water is eliminated during the reaction (Fig. 2.5). The amino acid units in a peptide chain are referred to as amino acid residues. A peptide chain consisting of three amino acid residues is called a tripeptide, e.g. glutathione in Figure 2.6. By convention, the amino terminus (N-terminus) is taken as the first residue, and the sequence of amino acids is written from left to right. When writing the peptide sequence, one uses either the three-letter or the one-letter abbreviations of amino acids, such as Asp-Arg-Val-Tyr-Ile-His-Pro-Phe-His-Leu or D-R-V-Y-I-H-P-F-H-L (see Table 2.1). This peptide is angiotensin, a peptide hormone that affects blood pressure. The amino acid residue having a free amino group at one end of the peptide, Asp, is called the N-terminal amino acid (amino terminus), whereas the residue having a free carboxyl group at the other end, Leu, is called the C-terminal amino acid (carboxyl terminus). Proteins contain between 50 and 2000 amino acid residues. The mean molecular mass of an amino acid residue is about 110 dalton units (Da). Therefore the molecular mass of most proteins is between 5500 and 220,000 Da. Human carbonic anhydrase I, an enzyme that plays a major role in acid–base balance in blood (Chapter 24), is a protein with a molecular mass of 29,000 Da (29 kDa).
Amino acids side chains contribute both charge and hydrophobicity to proteins
The amino acid composition of a peptide chain has a profound effect on its physical and chemical properties. Proteins rich in aliphatic or aromatic amino groups are relatively insoluble in water and are likely to be found in cell membranes. Proteins rich in polar amino acids are more water soluble. Amides are neutral compounds so that the amide backbone of a protein, including the α-amino and α-carboxyl groups from which it is formed, does not contribute to the charge of the protein. Instead, the charge on the protein is dependent on the side chain functional groups of amino acids. Amino acids with side chain acidic (Glu, Asp) or basic (Lys, His, Arg) groups will confer charge and buffering capacity to a protein. The balance between acidic and basic side chains in a protein determines its isoelectric point (pI) and net charge in solution. Proteins rich in lysine and arginine are basic in solution and have a positive charge at neutral pH, while acidic proteins, rich in aspartate and glutamate, are acidic and have a negative charge. Because of their side chain functional groups, all proteins become more positively charged at acidic pH and more negatively charged at basic pH. Proteins are an important part of the buffering capacity of cells and biological fluids, including blood.
Secondary structure of proteins
The secondary structure of a protein is determined by hydrogen bonding interactions between amino acid side chain functional groups
The secondary structure of a protein refers to the local structure of the polypeptide chain. This structure is determined by hydrogen bond interactions between the carbonyl oxygen group of one peptide bond and the amide hydrogen of another nearby peptide bond. There are two types of secondary structure: the α-helix and the β-pleated sheet.
The α-helix
The α-helix is a rod-like structure with the peptide chain tightly coiled and the side chains of amino acid residues extending outward from the axis of the spiral. Each amide carbonyl group is hydrogen-bonded to the amide hydrogen of a peptide bond that is four residues away along the same chain. There are on average 3.6 amino acid residues per turn of the helix, and the helix winds in a right-handed (clockwise) manner in almost all natural proteins (Fig. 2.7A).
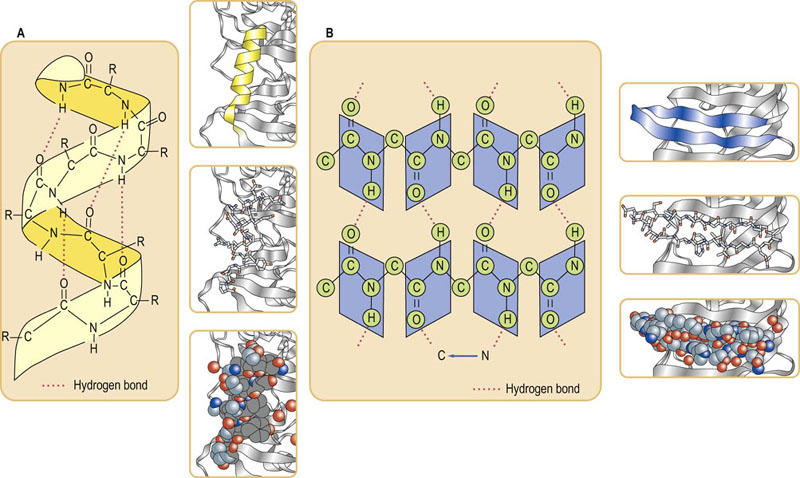
Fig. 2.7 Protein secondary structural motifs.
(A) An α-helical secondary structure. Hydrogen bonds between ‘backbone’ amide NH and C O groups stabilize the α-helix. Hydrogen atoms of OH, NH or SH group (hydrogen donors) interact with electron pairs of the acceptor atoms such as O, N or S. Even though the bonding energy is lower than that of covalent bonds, hydrogen bonds play a pivotal role in the stabilization of protein molecules. R, side chain of amino acids which extend outward from the helix. Ribbon, stick and space-filling models are shown. (B) The parallel β-sheet secondary structure. In the β-conformation, the backbone of the polypeptide chain is extended into a zigzag structure. When the zigzag polypeptide chains are arranged side by side, they form a structure resembling a series of pleats. Ribbon, stick and space-filling models are also shown.
O groups stabilize the α-helix. Hydrogen atoms of OH, NH or SH group (hydrogen donors) interact with electron pairs of the acceptor atoms such as O, N or S. Even though the bonding energy is lower than that of covalent bonds, hydrogen bonds play a pivotal role in the stabilization of protein molecules. R, side chain of amino acids which extend outward from the helix. Ribbon, stick and space-filling models are shown. (B) The parallel β-sheet secondary structure. In the β-conformation, the backbone of the polypeptide chain is extended into a zigzag structure. When the zigzag polypeptide chains are arranged side by side, they form a structure resembling a series of pleats. Ribbon, stick and space-filling models are also shown.
The β-pleated sheet
If the H-bonds are formed laterally between peptide bonds, the polypeptide sequences become arrayed parallel or antiparallel to one another in what is commonly called a β-pleated sheet. The β-pleated sheet is an extended structure as opposed to the coiled α-helix. It is pleated because the carbon–carbon (C C) bonds are tetrahedral and cannot exist in a planar configuration. If the polypeptide chain runs in the same direction, it forms a parallel β-sheet (Fig. 2.7B), but in the opposite direction, it forms an antiparallel structure. The β-turn or β-bend refers to the segment in which the polypeptide abruptly reverses direction. Glycine (Gly) and proline (Pro) residues often occur in β-turns on the surface of globular proteins.
C) bonds are tetrahedral and cannot exist in a planar configuration. If the polypeptide chain runs in the same direction, it forms a parallel β-sheet (Fig. 2.7B), but in the opposite direction, it forms an antiparallel structure. The β-turn or β-bend refers to the segment in which the polypeptide abruptly reverses direction. Glycine (Gly) and proline (Pro) residues often occur in β-turns on the surface of globular proteins.
Tertiary structure of proteins
The tertiary structure of a protein is determined by interactions between side chain functional groups, including disulfide bonds, hydrogen bonds, salt bridges, and hydrophobic interactions
The three-dimensional, folded and biologically active conformation of a protein is referred to as its tertiary structure. This structure reflects the overall shape of the molecule and generally consists of several smaller folded units termed domains. The tertiary structure of proteins is determined by X-ray crystallography and nuclear magnetic resonance spectroscopy.
The tertiary structure of a protein is stabilized by interactions between side chain functional groups: covalent disulfide bonds, hydrogen bonds, salt bridges, and hydrophobic interactions (Fig. 2.8). The side chains of tryptophan and arginine serve as hydrogen donors, whereas asparagine, glutamine, serine, and threonine can serve as both hydrogen donors and acceptors. Lysine, aspartic acid, glutamic acid, tyrosine, and histidine also can serve as both donors and acceptors in the formation of ion pairs (salt bridges). Two opposite-charged amino acids, such as glutamate with a γ-carboxyl group and lysine with an ε-amino group, may form a salt bridge, primarily on the surface of proteins (see Fig. 2.8).
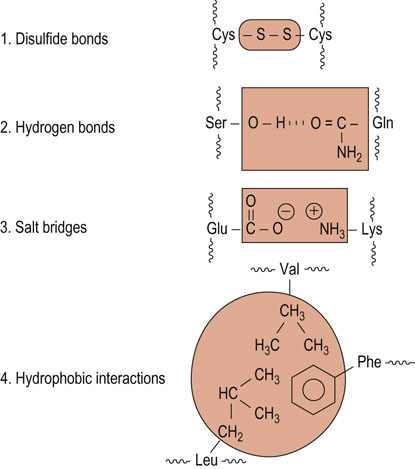
Fig. 2.8 Elements of tertiary structure of proteins.
Examples of amino acid side-chain interactions contributing to tertiary structure.
Compounds such as urea and guanidine hydrochloride cause denaturation or loss of secondary and tertiary structure when present at high concentrations for example, 8 mol/L urea. These reagents are called denaturants or chaotropic agents.
Quaternary structure of proteins is formed by interactions between peptide chains
The quaternary structure of multi-subunit proteins is determined by covalent and non-covalent interactions between the subunit surfaces
Quaternary structure refers to a complex or an assembly of two or more separate peptide chains that are held together by noncovalent or, in some cases, covalent interactions. In general, most proteins larger than 50 kDa consist of more than one chain and are referred to as dimeric, trimeric or multimeric proteins. Many multisubunit proteins are composed of different kinds of functional subunits, such as the regulatory and catalytic subunits. Hemoglobin is a tetrameric protein (Chapter 5), and beef heart mitochondrial ATPase has 10 protomers (Chapter 9). The smallest unit is referred to as a monomer or subunit. Figure 2.9 illustrates the structure of the dimeric protein Cu, Zn-superoxide dismutase. Figure 2.10 is an overview of the primary, secondary, tertiary, and quaternary structures of a tetrameric protein.
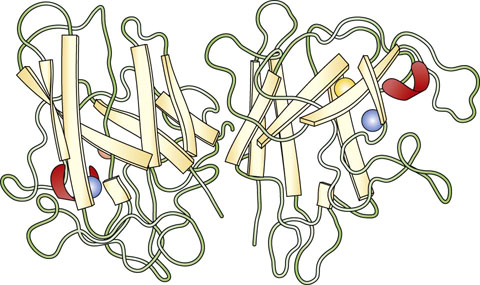
Fig. 2.9 Three-dimensional structure of a dimeric protein.
Quaternary structure of Cu,Zn-superoxide dismutase from spinach. Cu,Zn-superoxide dismutase has a dimeric structure, with a monomer molecular mass of 16,000 Da. Each subunit consists of eight antiparallel β-sheets called a β-barrel structure, in analogy with geometric motifs found on native American and Greek weaving and pottery. Red arc = intrachain disulfide bond. Courtesy of Dr Y. Kitagawa.
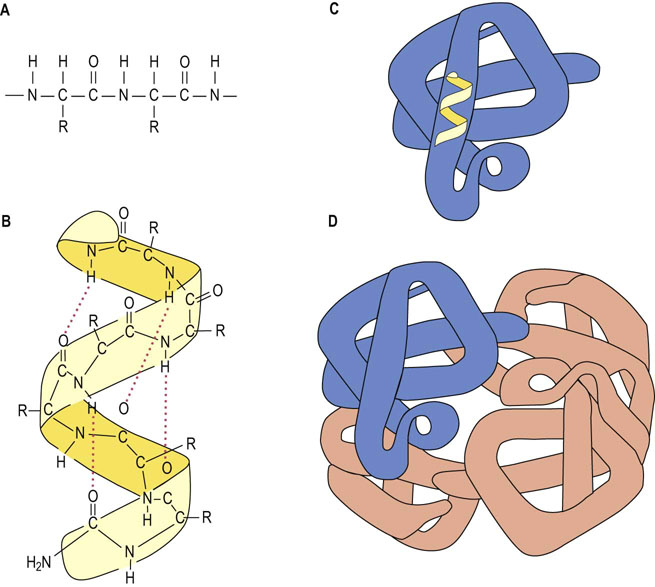
Fig. 2.10 Primary, secondary, tertiary, and quaternary structures.
(A) The primary structure is composed of a linear sequence of amino acid residues of proteins. (B) The secondary structure indicates the local spatial arrangement of polypeptide backbone yielding an extended α-helical or β-pleated sheet structure as depicted by the ribbon. Hydrogen bonds between the ‘backbone’ amide NH and C=O groups stabilize the helix. (C) The tertiary structure illustrates the three-dimensional conformation of a subunit of the protein, while the quaternary structure (D) indicates the assembly of multiple polypeptide chains into an intact, tetrameric protein.
Purification and characterization of proteins
Protein purification is a multi-step process, based on protein size, charge, solubility and ligand binding
Protein purification procedures take advantage of separations based on charge, size, binding properties, and solubility. The complete characterization of the protein requires an understanding of its amino acid composition, its complete primary, secondary and tertiary structure and, for multimeric proteins, their quaternary structure.
In order to characterize a protein, it is first necessary to purify the protein by separating it from other components in complex biological mixtures. The source of the proteins is commonly blood or tissues, or microbial cells such as bacteria and yeast. First, the cells or tissues are disrupted by grinding or homogenization in buffered isotonic solutions, commonly at physiologic pH and at 4°C to minimize protein denaturation during purification. The ‘crude extract’ containing organelles such as nuclei, mitochondria, lysosomes, microsomes, and cytosolic fractions can then be fractionated by high-speed centrifugation or ultracentrifugation. Proteins that are tightly bound to the other biomolecules or membranes may be solubilized using organic solvent or detergent.
Salting out (ammonium sulfate fractionation) and adjustment of pH
The solubility of a protein is dependent on the concentration of dissolved salts
The solubility of a protein may be increased by the addition of salt at a low concentration (salting in) or decreased by high salt concentration (salting out). When ammonium sulfate, one of the most soluble salts, is added to a solution of a protein, some proteins precipitate at a given salt concentration while others do not. Human serum immunoglobulins are precipitable by 33–40% saturated (NH4)2SO4, while albumin remains soluble. Saturated ammonium sulfate is about 4.1 mol/L. Most proteins will precipitate from an 80% saturated (NH4)2SO4 solution.
Proteins may also be precipitated from solution by adjusting the pH. Proteins are generally least soluble at their isoelectric point (pI). At this pH, the protein has no net charge or charge-charge repulsion between subunits. Hydrophobic interactions between protein surfaces may lead to aggregation and precipitation of the protein.
Separation on the basis of size
Dialysis and ultrafiltration
Small molecules, such as salts, can be removed from protein solutions by dialysis or ultrafiltration
Dialysis is performed by adding the protein–salt solution to a semipermeable membrane tube (commonly a nitrocellulose or collodion membrane). When the tube is immersed in a dilute buffer solution, small molecules will pass through and large protein molecules will be retained in the tube, depending on the pore size of the dialysis membrane. This procedure is particularly useful for removal of (NH4)2SO4 or other salts during protein purification, since the salts will interfere with the purification of proteins by ion exchange chromatography (below). Figure 2.11 illustrates the dialysis of proteins.
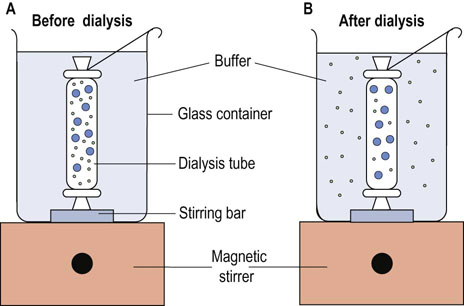
Fig. 2.11 Dialysis of proteins.
Protein and low-molecular-mass compounds are separated by dialysis on the basis of size. (A) A protein solution with salts is placed in a dialysis tube in a beaker and dialyzed with stirring against an appropriate buffer. (B) The protein is retained in the dialysis tube, whereas salts will exchange through the membrane. By use of a large volume of external buffer, with occasional buffer replacement, the protein will eventually be exchanged into the external buffer solution.
Ultrafiltration has largely replaced dialysis for purification of proteins. This technique uses pressure to force a solution through a semipermeable membrane of defined, homogeneous pore size. By selecting the proper molecular weight cut-off value (pore size) for the filter, the membranes will allow solvent and lower molecular weight solutes to permeate the membrane, forming the filtrate, while retaining higher molecular weight proteins in the retentate solution. Ultrafiltration can be used to concentrate protein solutions or to accomplish dialysis by continuous replacement of buffer in the retentate compartment.
Gel filtration (molecular sieving)
Gel filtration chromatography separates proteins on the basis of size
Gel filtration, or gel permeation, chromatography uses a column of insoluble but highly hydrated polymers such as dextrans, agarose or polyacrylamide. Gel filtration chromatography depends on the differential migration of dissolved solutes through gels that have pores of defined sizes. This technique is frequently used for protein purification and for desalting protein solutions. Figure 2.12 describes the principle of gel filtration. There are commercially available gels made from carbohydrate polymer beads designated as dextran (Sephadex series), polyacrylamide (Bio-Gel P series), and agarose (Sepharose series), respectively. The gels vary in pore size and one can choose the gel filtration materials according to the molecular weight fractionation range desired.
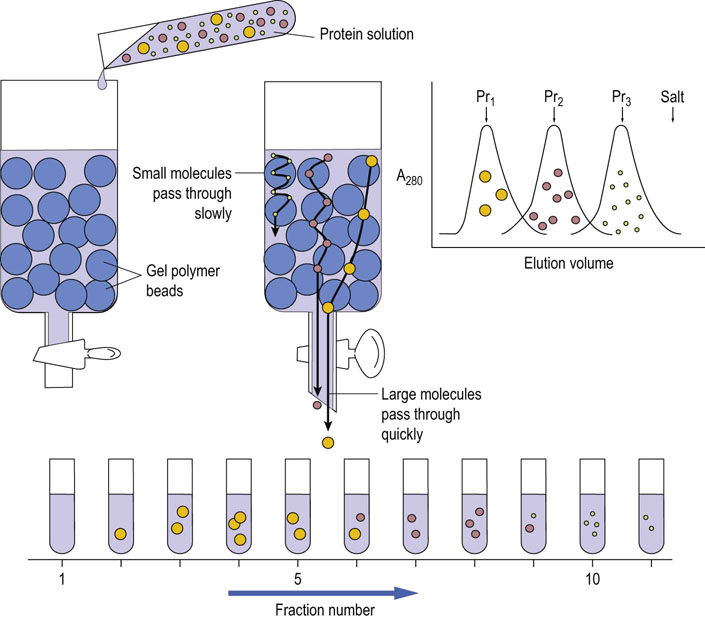
Fig. 2.12 Fractionation of proteins by size: gel filtration chromatography of proteins.
Proteins with different molecular sizes are separated by gel filtration based on their relative size. The smaller the protein, the more readily it exchanges into polymer beads, whereas larger proteins may be completely excluded. Larger molecules flow more rapidly through this column, leading to fractionation on the basis of molecular size. The chromatogram on the right shows a theoretical fractionation of three proteins, Pr1–Pr3 of decreasing molecular weight.
Ion exchange chromatography
Proteins bind to ion exchange matrices based on charge-charge interactions
When a charged ion or molecule with one or more positive charges exchanges with another positively charged component bound to a negatively charged immobilized phase, the process is called cation exchange. The inverse process is called anion exchange. The cation exchanger, carboxymethylcellulose ( O
O CH2
CH2 COO
COO ), and anion exchanger, diethylaminoethyl (DEAE) cellulose [
), and anion exchanger, diethylaminoethyl (DEAE) cellulose [ O
O C2H4
C2H4 NH+(C2H5)2], are frequently used for the purification of proteins. Consider purifying a protein mixture containing albumin and immunoglobulin. At pH 7.5, albumin, with a pI of 4.8, is negatively charged; immunoglobulin with a pI ∼8 is positively charged. If the mixture is applied to a DEAE column at pH 7, the albumin sticks to the positively charged DEAE column whereas the immunoglobulin passes through the column. Figure 2.13 illustrates the principle of ion exchange chromatography. As with gel permeation chromatography, proteins can be separated from one another, based on small differences in their pI. Adsorbed proteins are commonly eluted with a gradient formed from two or more solutions with different pH and/or salt concentrations. In this way, proteins are gradually eluted from the column and are well resolved based on their pI.
NH+(C2H5)2], are frequently used for the purification of proteins. Consider purifying a protein mixture containing albumin and immunoglobulin. At pH 7.5, albumin, with a pI of 4.8, is negatively charged; immunoglobulin with a pI ∼8 is positively charged. If the mixture is applied to a DEAE column at pH 7, the albumin sticks to the positively charged DEAE column whereas the immunoglobulin passes through the column. Figure 2.13 illustrates the principle of ion exchange chromatography. As with gel permeation chromatography, proteins can be separated from one another, based on small differences in their pI. Adsorbed proteins are commonly eluted with a gradient formed from two or more solutions with different pH and/or salt concentrations. In this way, proteins are gradually eluted from the column and are well resolved based on their pI.
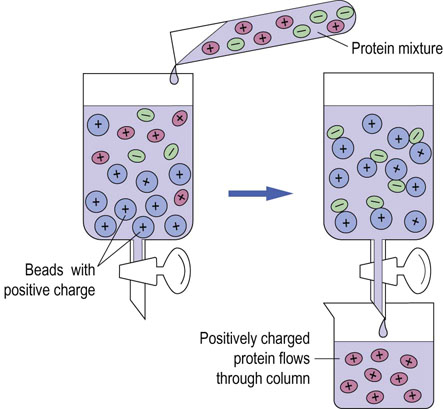
Fig. 2.13 Fractionation of proteins by charge: ion exchange chromatography.
Mixtures of proteins can be separated by ion exchange chromatography according to their net charges. Beads that have positively charged groups attached are called anion exchangers, whereas those having negatively charged groups are cation exchangers. This figure depicts an anion exchange column. Negatively charged protein binds to positively charged beads, and positively charged protein flows through the column.
Affinity chromatography
Affinity chromatography purifies proteins based on ligand interactions
Affinity chromatography is a convenient and specific method for purification of proteins. A porous chromatography column matrix is derivatized with a ligand that interacts with, or binds to, a specific protein in a complex mixture. The protein of interest will be selectively and specifically bound to the ligand while the others wash through the column. The bound protein can then be eluted by a high salt concentration, mild denaturation or by a soluble form of the ligand or ligand analogs (see Chapter 6).
Determination of purity and molecular weight of proteins
Polyacrylamide gel electrophoresis in sodium dodecylsulfate can used to separate proteins, based on charge
Electrophoresis can be used for the separation of a wide variety of charged molecules, including amino acids, polypeptides, proteins, and DNA. When a current is applied to molecules in dilute buffers, those with a net negative charge at the selected pH migrate toward the anode and those with a net positive charge toward the cathode. A porous support, such as paper, cellulose acetate or polymeric gel, is commonly used to minimize diffusion and convection.
Like chromatography, electrophoresis may be used for preparative fractionation of proteins at physiologic pH. Different soluble proteins will move at different rates in the electrical field, depending on their charge-to-mass ratio. A denaturing detergent, sodium dodecyl sulfate (SDS), is commonly used in a polyacrylamide gel electrophoresis (PAGE) system to separate and resolve protein subunits according to molecular weight. The protein preparation is usually treated with both SDS and a thiol reagent, such as β-mercaptoethanol, to reduce disulfide bonds. Because the binding of SDS is proportional to the length of the peptide chain, each protein molecule has the same mass-to-charge ratio and the relative mobility of the protein is proportional to the molecular mass of the polypeptide chain. Varying the state of crosslinking of the polyacrylamide gel provides selectivity for proteins of different molecular weights. A purified protein preparation can be readily analyzed for homogeneity on SDS-PAGE by staining with sensitive and specific dyes, such as Coomassie Blue, or with a silver staining technique, as shown in Figure 2.14.
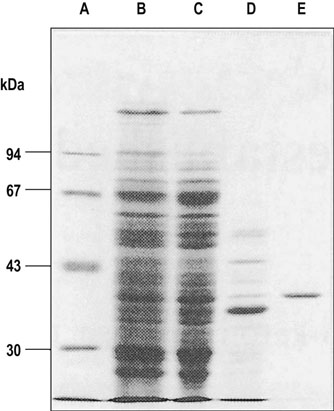
Fig. 2.14 SDS-PAGE.
Sodium dodecylsulfate-polyacrylamide gel electrophoresis is used to separate proteins on the basis of their molecular weights. Larger molecules are retarded in the gel matrix, whereas the smaller ones move more rapidly. Lane A contains standard proteins with known molecular masses (indicated in kDa on the left). Lanes B, C, D, and E show results of SDS-PAGE analysis of a protein at various stages in purification: B = total protein isolate; C = ammonium sulfate precipitate; D = fraction from gel permeation chromatography; E = purified protein from ion exchange chromatography.
Isoelectric focusing (IEF)
Isoelectric focusing resolves proteins based on their isoelectric point
Isoelectric focusing (IEF) is conducted in a microchannel or gel containing a stabilized pH gradient. A protein applied to the system will be either positively or negatively charged, depending on its amino acid composition and the ambient pH. Upon application of a current, the protein will move towards either the anode or cathode until it encounters that part of the system which corresponds to its pI, where the protein has no charge and will cease to migrate. IEF is used in conjunction with SDS-PAGE for two-dimensional gel electrophoresis (Fig. 2.15). This technique is particularly useful for the fractionation of complex mixtures of proteins for proteomic analysis.
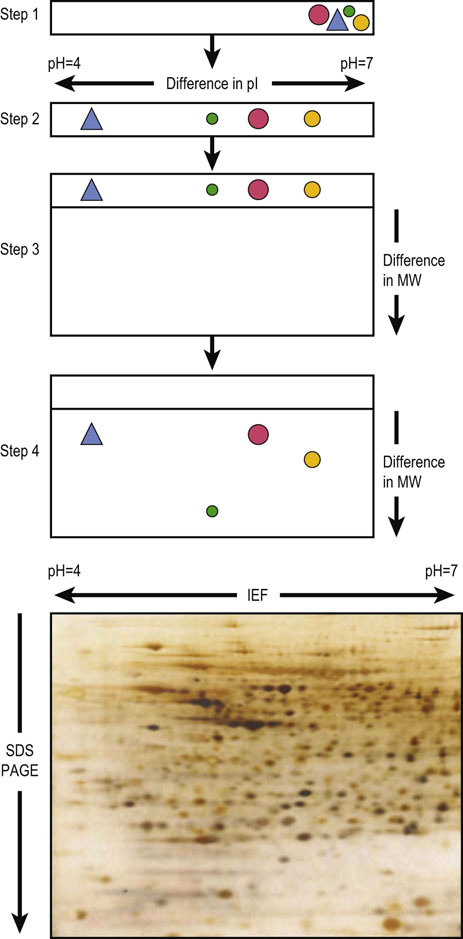
Fig. 2.15 Two-dimensional gel electrophoresis.
(top) Step 1: Sample containing proteins is applied to a cylindrical isoelectric focusing gel within the pH gradient. Step 2: Each protein migrates to a position in the gel corresponding to its isoelectric point (pI). Step 3: The IEF gel is placed horizontally on the top of a slab gel. Step 4: The proteins are separated by SDS-PAGE according to their molecular weight. (bottom) Typical example of 2D-PAGE. A rat liver homogenate was fractionated by 2D-PAGE and proteins were detected by silver staining.
Analysis of protein structure
The typical steps in the purification of a protein are summarized in Figure 2.16. Once purified, for the determination of its amino acid composition, a protein is subjected to hydrolysis, commonly in 6 mol/L HCl at 110°C in a sealed and evacuated tube for 24–48 h. Under these conditions, tryptophan, cysteine and most of the cystine are destroyed, and glutamine and asparagine are quantitatively deaminated to give glutamate and aspartate, respectively. Recovery of serine and threonine is incomplete and decreases with increasing time of hydrolysis.
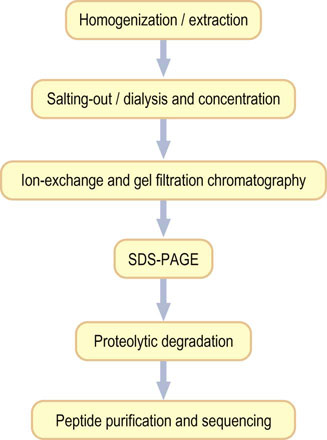
Fig. 2.16 Strategy for protein purification.
Purification of a protein involves a sequence of steps in which contaminating proteins are removed, based on difference in size, charge, and hydrophobicity. Purification is monitored by SDS-PAGE (see Fig. 2.14). The primary sequence of the protein may be determined by automated Edman degradation of peptides (see Fig. 2.18). The three-dimensional structure of the protein may be determined by X-ray crystallography.
Alternative hydrolysis procedures may be used for measurement of tryptophan, while cysteine and cystine may be converted to an acid-stable cysteic acid prior to hydrolysis. Following hydrolysis, the free amino acids are separated on an automated amino acid analyzer using an ion exchange column or, following pre-column derivatization with colored or fluorescent reagents, by reversed-phase high-performance liquid chromatography (HPLC). The free amino acids fractionated by ion exchange chromatography are detected by reaction with a chromogenic or fluorogenic reagent, such as ninhydrin or dansyl chloride, Edman's reagent (see below) or o-phthalaldehyde. These techniques allow the measurement of as little as 1 pmol of each amino acid. A typical elution pattern of amino acids in a purified protein is shown in Figure 2.17.
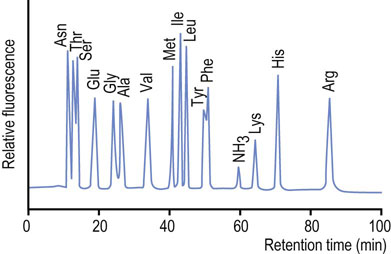
Fig. 2.17 Chromatogram from an amino acid analysis by cation-exchange chromatography.
A protein hydrolysate is applied to the cation exchange column in a dilute buffer at acidic pH (~3.0), at which all amino acids are positively charged. The amino acids are then eluted by a gradient of increasing pH and salt concentrations. The most anionic (acidic) amino acids elute first, followed by the neutral and basic amino acids. Amino acids are derived by post-column reaction with a fluorogenic compound, such as o-phthalaldehyde.
Determination of the primary structure of proteins
Historically, analysis of protein sequence was carried out by chemical methods; today, both sequence analysis and protein identification are performed by mass spectrometry
Information on the primary sequence of a protein is essential for understanding its functional properties, the identification of the family to which the protein belongs, as well as characterization of mutant proteins that cause disease. A protein may be cleaved first by digestion by specific endoproteases, such as trypsin (Chapter 6), V8 protease or lysyl endopeptidase, to obtain peptide fragments. Trypsin cleaves peptide bonds on the C-terminal side of arginine and lysine residues, provided the next residue is not proline. Lysyl endopeptidase is also frequently used to cleave at the C-terminal side of lysine. Cleavage by chemical reagents such as cyanogen bromide is also useful. Cyanogen bromide cleaves on the C-terminal side of methionine residues. Before cleavage, proteins with cysteine and cystine residues are reduced by 2-mercaptoethanol and then treated with iodoacetate to form carboxymethylcysteine residues. This avoids spontaneous formation of inter- or intramolecular disulfides during analyses.
The cleaved peptides are then subjected to reverse-phase HPLC to purify the peptide fragments, and then sequenced on an automated protein sequencer, using the Edman degradation technique (Fig. 2.18). The sequence of overlapping peptides is then used to obtain the primary structure of the protein. The Edman degradation technique is largely of historical interest. Mass spectrometry is more commonly used today to obtain both the molecular mass and sequence of polypeptides simultaneously (Chapter 36). Both techniques can be applied directly to proteins or peptides recovered from SDS-PAGE or two-dimensional electrophoresis (IEF plus SDS-PAGE).
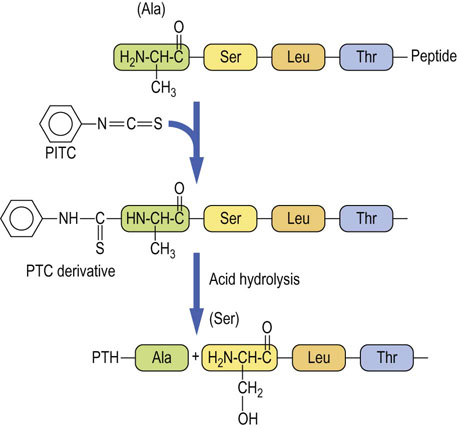
Fig. 2.18 Steps in Edman degradation.
The Edman degradation method sequentially removes one residue at a time from the amino end of a peptide. Phenyl isothiocyanate (PITC) converts the N-terminal amino group of the immobilized peptide to a phenylthiocarbamyl derivative (PTC amino acid) in alkaline solution. Acid treatment removes the first amino acid as the phenylthiohydantoin (PTH) derivative, which is identified by HPLC.
Protein sequencing and identification can also be done by electrospray ionization liquid chromatography tandem mass spectrometry (HPLC-ESI-MS/MS) (Chapter 36). This technique is sufficiently sensitive that proteins separated by 2D-PAGE (see Fig. 2.15) can be recovered from the gel for analysis. As little as 1 µg of protein per spot, can be digested with trypsin in situ, then extracted from the gel and identified, based on their amino acid sequence. This technique, as well as a complementary technique called matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) MS/MS (Chapter 36), can be applied for determination of the molecular weight of intact proteins, as well as for sequence analysis of peptides, leading to unambiguous identification of a protein.
Determination of the three-dimensional structure of proteins
X-ray crystallography and NMR spectroscopy are usually used for determination of the three-dimensional structure of proteins
X-ray crystallography depends on the diffraction of X-rays by the electrons of the atoms constituting the molecule. However, since the X-ray diffraction caused by an individual molecule is weak, the protein must exist in the form of a well-ordered crystal, in which each molecule has the same conformation in a specific position and orientation on a three-dimensional lattice. Based on diffraction of a collimated beam of electrons, the distribution of the electron density, and thus the location of atoms, in the crystal can be calculated to determine the structure of the protein. For protein crystallization, the most frequently used method is the hanging drop method which involves the use of a simple apparatus that permits a small portion of a protein solution (typically 10 µL droplet containing 0.5–1 mg/protein) to evaporate gradually to reach the saturating point at which the protein begins to crystallize. NMR spectroscopy is usually used for structural analysis of small organic compounds, but high-field NMR is also useful for determination of the structure of a protein in solution and complements information obtained by X-ray crystallography.
Summary
 A total of 20 alpha-amino acids are the building blocks of proteins. The side chains of these amino acids contribute charge, polarity and hydrophobicity to protein.
A total of 20 alpha-amino acids are the building blocks of proteins. The side chains of these amino acids contribute charge, polarity and hydrophobicity to protein.
 Proteins are macromolecules formed by polymerization of L-α-amino acids by peptide bonds. The linear sequence of the amino acids constitutes the primary structure of the protein.
Proteins are macromolecules formed by polymerization of L-α-amino acids by peptide bonds. The linear sequence of the amino acids constitutes the primary structure of the protein.
 Proteins are macromolecules formed by polymerization of L-α-amino acids. There are 20 different amino acids in proteins, linked by peptide bonds. The linear sequence of the amino acids is the primary structure of the protein.
Proteins are macromolecules formed by polymerization of L-α-amino acids. There are 20 different amino acids in proteins, linked by peptide bonds. The linear sequence of the amino acids is the primary structure of the protein.
 The higher-order structure of a protein is the product of its secondary, tertiary, and quaternary structure.
The higher-order structure of a protein is the product of its secondary, tertiary, and quaternary structure.
 These higher order structures are formed by hydrogen bonds, hydrophobic interactions, salt bridges and covalent bonds between the side chains of amino acids.
These higher order structures are formed by hydrogen bonds, hydrophobic interactions, salt bridges and covalent bonds between the side chains of amino acids.
 Purification and characterization of proteins are essential for elucidating their structure and function. By taking advantage of differences in their size, solubility, charge and ligand-binding properties, proteins can be purified to homogeneity using various chromatographic and electrophoretic techniques. The molecular mass and purity of a protein, and its subunit composition, can be determined by SDS-PAGE.
Purification and characterization of proteins are essential for elucidating their structure and function. By taking advantage of differences in their size, solubility, charge and ligand-binding properties, proteins can be purified to homogeneity using various chromatographic and electrophoretic techniques. The molecular mass and purity of a protein, and its subunit composition, can be determined by SDS-PAGE.
 Deciphering the primary and three-dimensional structures of a protein by chemical methods, mass spectrometry, X-ray analysis and NMR spectroscopy leads to an understanding of structure–function relationships in proteins.
Deciphering the primary and three-dimensional structures of a protein by chemical methods, mass spectrometry, X-ray analysis and NMR spectroscopy leads to an understanding of structure–function relationships in proteins.
Aguzzi, A, Falsig, J. Prion propagation, toxicity and degradation. Nat Neurosci. 2012; 15:936–939.
Dominguez, DC, Lopes, R, Torres, ML. Proteomics: clinical applications. Clin Lab Sci. 2007; 20:245–248.
Griffin, MD, Gerrard, JA. The relationship between oligomeric state and protein function. Adv Exp Med Biol. 2012; 747:74–90.
Kovacs, GG, Budka, H. Prion diseases: from protein to cell pathology. Am J Pathol. 2008; 172:555–565.
Marouga, R, David, S, Hawkins, E. The development of the DIGE system: 2D fluorescence difference gel analysis technology. Anal Bioanal Chem. 2005; 382:669–678.
Matt, P, Fu, Z, Ru, Q, Van Eyk, JE. Biomarker discovery: proteome fractionation and separation in biological samples. J Physiol Genomics. 2008; 14:12–17.
Shkundina, IS, Ter-Avanesyan, MD. Prions. Biochemistry (Moscow). 2007; 72:1519–1536.
Sułkowska, JI, Rawdon, EJ, Millett, KC, et al. Conservation of complex knotting and slipknotting patterns in proteins. Proc Natl Acad Sci U S A. 2012; 109:E1715–1723.
Walsh, CT. Posttranslational modification of proteins: expanding nature's inventory, ed 3. Colorado: Roberts & Co.; 2007.
Protein Data Ban. www.rcsb.org [– Use Search Box, then select a structure and view protein in Jmol].
www.ncbi.nlm.nih.gov/Structure [– National Center for Biotechnology Information, National Library of Medicine. Several databases, including protein structure].
http://us.expasy.org [– Bioinformatics resource portal].